Figure 1.
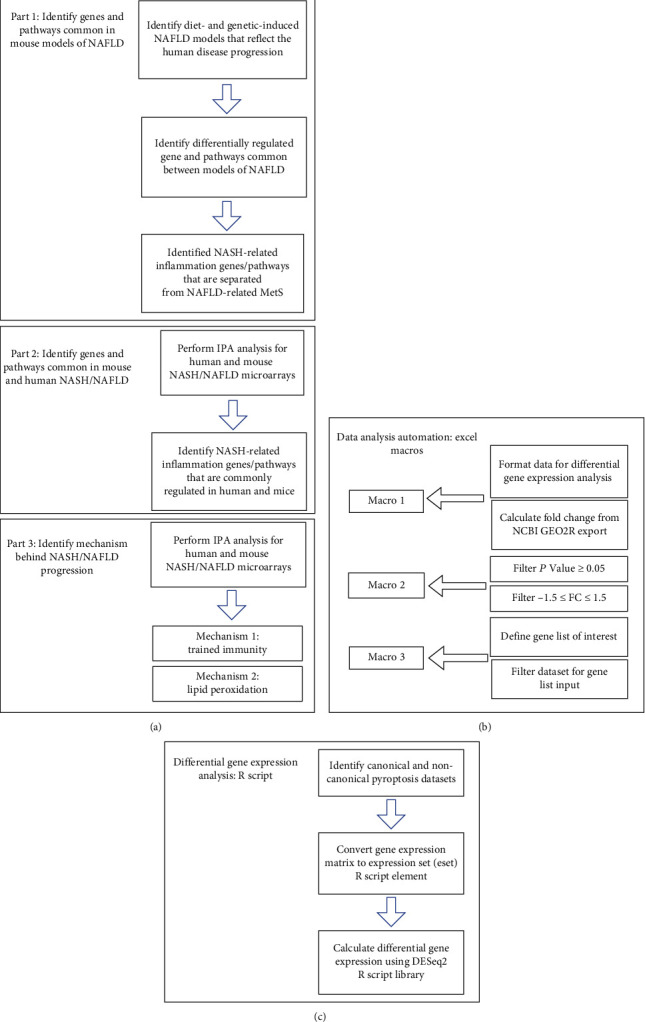
The flowchart of our data mining analyses. (a) Workflow included three parts: (1) identify genes and signaling pathways shared by microarrays from several mouse models of nonalcoholic fatty liver disease (NAFLD), (2) identify genes and signaling pathways common in mouse models of NAFLD and patients with nonalcoholic steatohepatitis (NASH), and (3) identify significant inflammatory mechanisms underlying the pathogenesis and progression of NASH/NAFLD. (b) Process automations using Microsoft Excel Macros, which have significantly facilitated the database mining processes comparing to that of our previous database mining papers. (c) GEO datasets without the GEO2R function were analyzed using DESeq2 library in R Studio. R script code used in C: Meeta Mistry, C. Titus Brown, jessicalumian, & tug65470 (2021, July 14), tug65470/msu_ngs2015: DESeq2 analysis of microarray and RNA-seq datasets (version v1.0.0), Zenodo, http://doi.org/10.5281/zenodo.5102949.
