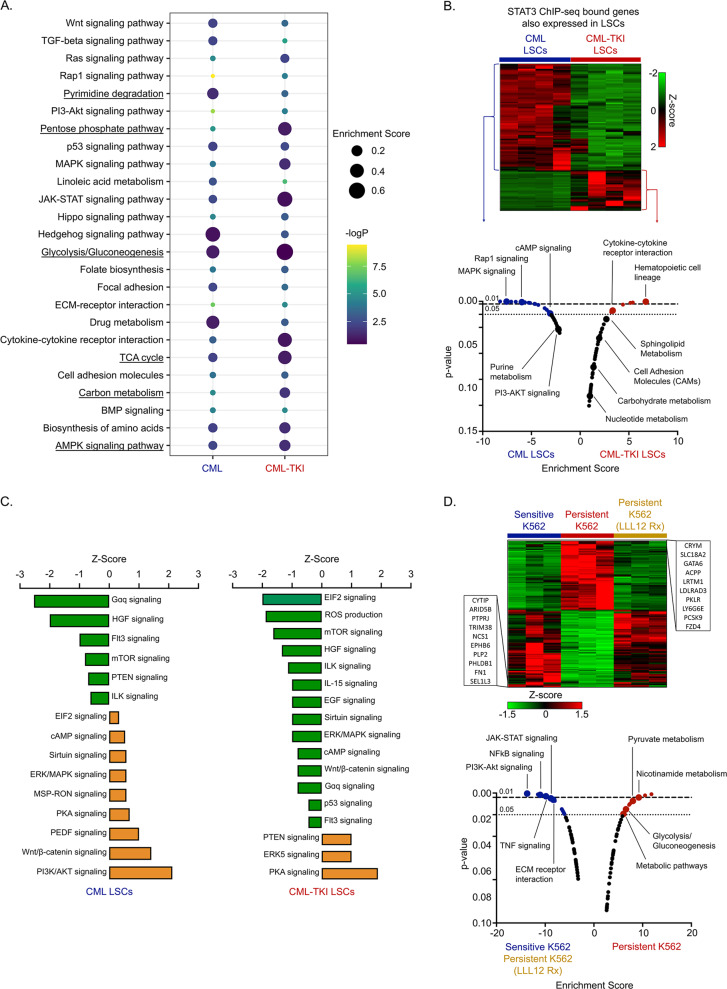
Fig. 3. STAT3 localizes to metabolic genes in transcriptionally altered TKI-persistent LSCs.
A Kegg GO Pathway analysis depicting z score and p value for the genes differentially bound by pSTAT3-Y705 in the CML and IM-persistent CML LSK. B Heatmap and pathway analysis for integration of the pSTAT3-Y705 bound ChIP-seq peaks and the upregulated RNA-seq genes in the CML and IM-persistent LSK and LSCs respectively. C Ingenuity Pathway analysis bar chart depicting z scores of pathways correlated to overlapping genes of CML vs WT bulk-RNA sequencing and pSTAT3-Y705 ChIP-seq peaks unique to CML as well as overlapping genes of CML-TKI vs WT bulk-RNA sequencing and pSTAT3-Y705 ChIP-seq peaks unique to CML-TKI. D Heatmap and pathway analysis for upregulated genes common between sensitive K562 and IM-persistent K562 treated with 1 µM LLL12 (1 µM) but different from IM-persistent K562. The transcriptomic data were carried out in triplicates.