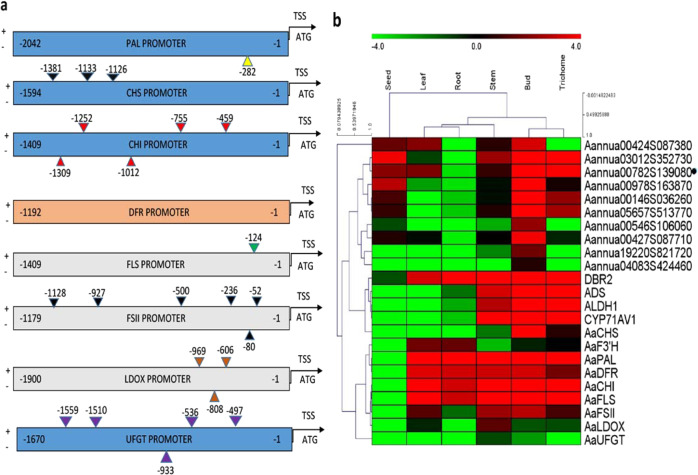
Fig. 2. Bioinformatic analysis of the promoters of flavonoid biosynthetic genes, as well as the expression profile of YABBY family genes and flavonoid-regulating genes in A. annua.
a Putative YABBY-binding sites predicted by PLANTPAN3.0 are shown. Positions on plus and minus strands are represented by numbers above and below the promoter sequence, respectively. YABBY-binding sites were found in all promoters except the DFR promoter sequence, for which no predicted YABBY-binding site was predicted. b Heatmap showing the expression profile of YABBY family genes, as well as flavonoid biosynthetic genes in six tissues. The color scale at the top represents the RPKM (reads per kilobase per million mapped reads) values. AaYABBY5 (marked with a black dot) was selected as a potential transcription factor that might regulate flavonoid biosynthesis because of its similar expression pattern to the important flavonoid biosynthetic genes