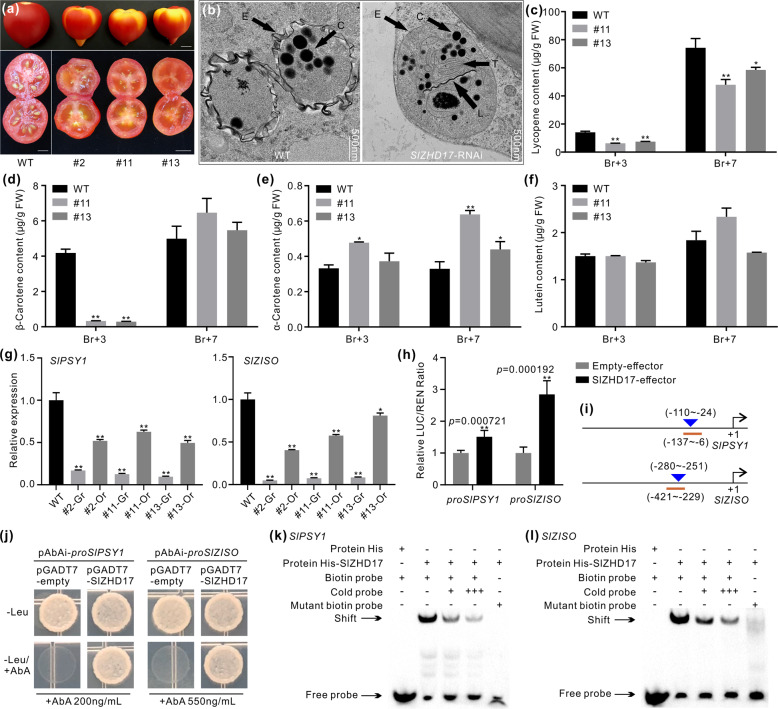
Fig. 6. SlZHD17 influences carotenoid accumulation by directly regulating the SlPSY1 and SlZISO genes.
a Photograph of the whole and cross sections of WT and SlZHD17-RNAi fruit at the Br+7 stage. Bar = 5 mm. b Ultrastructure of chromoplasts in the mesocarp of WT and SlZHD17-RNAi (line #11) Br+3 fruit observed by TEM. C carotenoid-containing globules, E plastid envelope, L crystal line, and T thylakoid grana. c–f The content of lycopene (c), β-carotene (d), α-carotene (e), and lutein (f) in WT and SlZHD17-RNAi fruit at the Br+3 and Br+7 stages by HPLC. FW, fresh weight. g The relative expression levels of SlPSY1 and SlZISO in WT and SlZHD17-RNAi Br+3 fruit. For each transgenic fruit, the normal orange part (Or) and abnormal green part (Gr) were sampled separately. In c–g, data represent the mean values of three independent experiments, and error bars show the standard error values. Single asterisk (*) and double asterisks (**) refer to significant differences between WT and transgenic lines with P < 0.05 and P < 0.01, respectively (two-tailed Student’s t-test). h Regulation of SlZHD17 on SlPSY1 and SlZISO gene promoters based on dual-luciferase assay. The empty effector was used as a control (set as 1). Data represent the mean values of six independent experiments, and error bars show the standard error values. Double asterisks (**) refers to significant differences between empty effector and SlZHD17-effector with P < 0.01, respectively (two-tailed Student’s t-test). The P values were provided in the graph. i Schematic diagrams of sequence positions chosen for yeast one-hybrid assay and electrophoretic mobility shift assay (EMSA). The underlined region indicates the promoter fragment used for the yeast one-hybrid assay, and the triangle indicates the sequence position used for the EMSA. The detailed sequences are provided in Appendix S4. j SlZHD17 binding with SlPSY1 and SlZISO promoter fragments assessed by yeast one-hybrid assay. The yeast transformants were cultured on SD/−Leu and SD/−Leu/+AbA media for 3–5 days, and the pGADT7-empty plasmid was used as a control. k, l SlZHD17 binding with SlPSY1 (k) and SlZISO (l) promoter regions in vitro by EMSA. TF-His protein incubated with a biotin-labeled DNA probe was used as a negative control. ‘+++’ indicates an increase in the competitor probe; no shift in the band for the His-SlZHD17 protein and mutant biotin-labeled DNA probe further confirmed the specific binding site