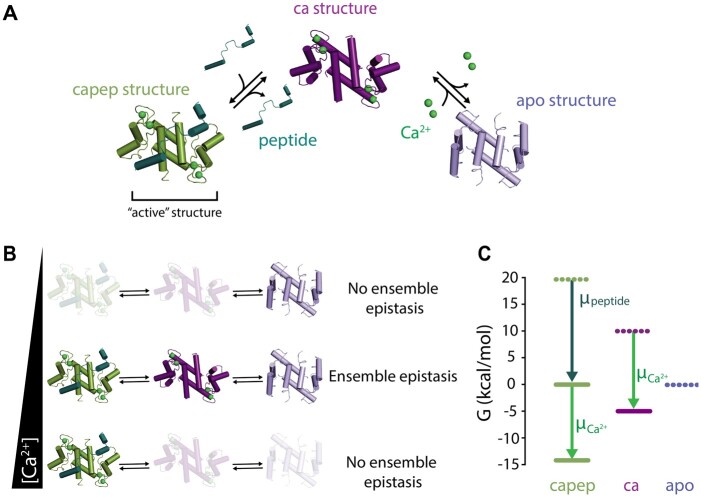
Figure 5.
Testing for ensemble epistasis in the S100A4 protein. (A) Three-conformation ensemble of the S100A4 protein. The apo conformation (apo, slate, PDB: 1M31) is in equilibrium with the bound (ca, purple, PDB: 2Q91) and /peptide bound (capep, green, PDB: 5LPU) conformations when (lime green spheres) and peptide (dark green) are present. (B) The relative populations of the apo, ca, and capep conformations change as concentration increases in the presence of saturating peptide. The magnitude of ensemble epistasis observed is -dependent, because only some concentrations lead to multiple populated conformations. (C) Assigned energies () of S100A4 conformations. Apo is most stable when peptide, μpeptide, and chemical potentials, , are zero (dashed lines). Capep is stabilized by increasing (dark green arrow, solid green line). Increasing alters the energies of both ca and capep (lime green arrow, solid lines). All calculations were done at .