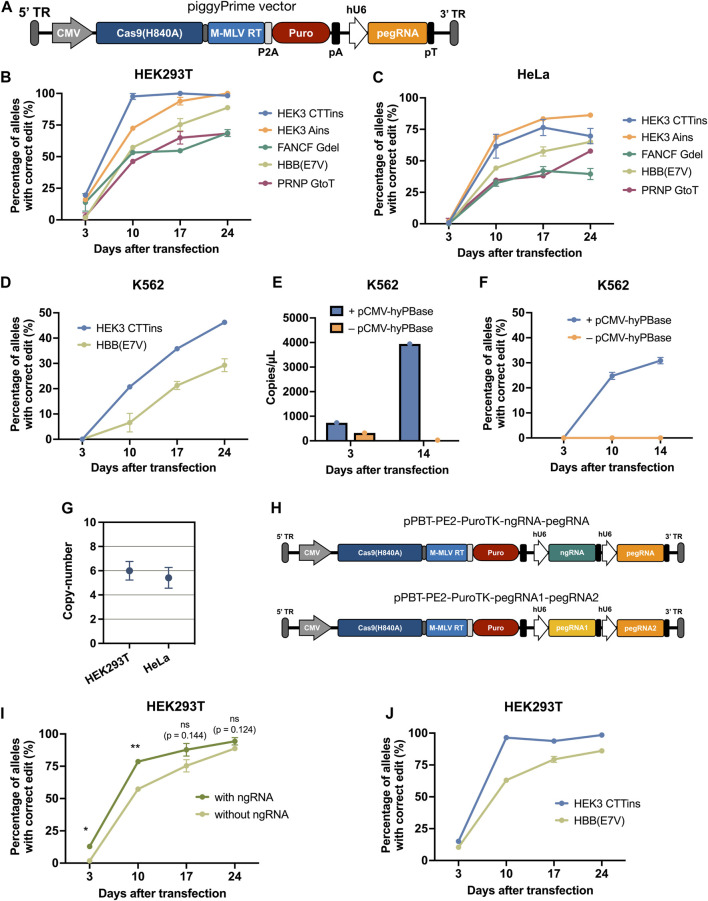
FIGURE 2.
| Effective prime editing by piggyBac-mediated integration of all prime editing components using a single-vector system. (A) Schematic overview of the piggyPrime vector, consisting of (from the right) a 5’ terminal repeat (TR), CMV promotor, Cas9(H840A)-linker-M-MLV-RT (PE2), P2A, PuroR, bGH pA, hU6 promotor, pegRNA, and 3’ TR. (B,C) Integration of piggyPrime vectors containing the HEK3-CTTins, HEK3-Ains, HBB(E7V), FANCF-6Gdel, and PRNP-GtoT pegRNAs into HEK293T (B) and HeLa (C) cells resulting in increasing correct editing at target sites over time. Puromycin was applied at day 3 after transfection. (D) piggyPrime vectors can also successfully be integrated into the genome of K562 cells by transfection, resulting in increased correct editing over time. (E) pegRNA levels were determined at day 3 and 14 in K562 cells transfected with the HEK3-CTTins piggyPrime vector either with or without hyPBase. Only cells co-transfected with the piggyPrime vector and hyPBase-encoding plasmid DNA showed detectable pegRNA levels at day 14. (F) Editing rates were determined at multiple time points in K562 cells transfected with the HEK3-CTTins piggyPrime vector either with or without hyPBase. Only cells co-transfected with the piggyPrime vector and hyPBase-encoding plasmid DNA showed detectable correct editing. (G) The average copy-number of HEK3-CTTins piggyPrime vectors was determined in HEK293T and HeLa cells using ddPCR. (H) Schematic overview of piggyPrime vectors carrying both a nicking sgRNA (ngRNA) and a pegRNA (top) or dual pegRNAs (bottom). (I) Integration of HBB(E7V) piggyPrime vector carrying both a ngRNA and a pegRNA cassette resulted in markedly increased editing compared to HBB(E7V) piggyPrime vectors without a ngRNA. (J) Integration of piggyPrime vectors carrying both the HEK3-CTTins and HBB(E7V) pegRNA resulted in correct editing at both target sites, without compromising editing efficacy. Data and error bars show mean (n = 3) ± sd. Statistical significance was calculated using multiple unpaired t-tests with correction for multiple testing (*p < 0.03, **p < 0.002).