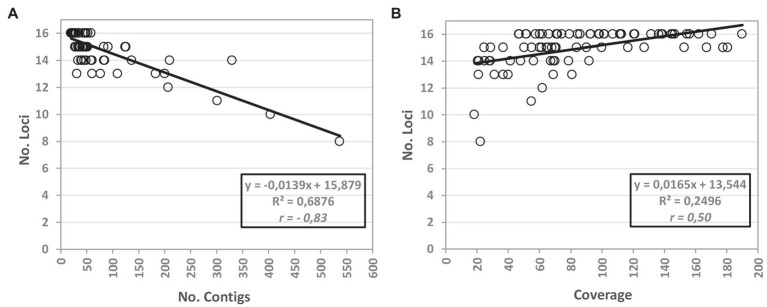
Figure 3.
Influence of the mean coverage depth and number of contigs on the efficacy of the WGS-based MLVA extraction. The graphs show the correlation of the efficacy (measured by the number of loci for which an allele was called) of the bioinformatics script with the number of assembled contigs (A) and with the depth of coverage (B) after quality improvement. For (A,B), the tendency lines are shown with the respective equations and the Pearson coefficient (r). The WGS-based results are relative to the MLVA loci extraction using draft genomes assembled with the INNUca v4.0.1 pipeline using Pilon v1.18 (Bankevich et al., 2012).