Figure 2.
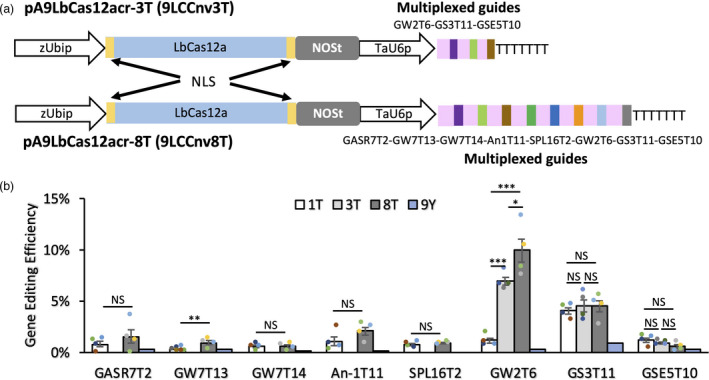
The gene editing efficiency of CRISPR‐LbCas12a constructs with single and multiple crRNA units. (a) Schematic illustration of the CRISPR‐LbCas12a MGE constructs. The MGE constructs targeting three and eight targets are referred to as pA9LbCas12acr‐3T (or 9LCCnv3T) and pA9LbCas12acr‐8T (or 9LCCnv8T), respectively. The order of guide sequences for different targets are shown next to the construct names. (b) The bar plots of gene editing efficiency for the CRISPR‐LbCas12a constructs targeting one (1T), three (3T) and eight (8T) targets. Data are shown as mean ± standard error. The data generated from the protoplasts transformed with construct pA9eYFP that carries the YFP gene was used as negative control (marked as 9Y). Because targets located in the A, B and D genomes showed similar LbCas12a‐induced mutation rates, data generated for all three genomes was pooled together for calculating the gene editing efficiency. The analyses of each target site were based on four biological replicates, the result of each replicate is shown as coloured dots on the bar plots. Student’s t test was used to assess the significance of differences in gene editing efficiency between guides from the constructs with one, three or eight crRNAs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; NS P > 0.05.
