Figure 4.
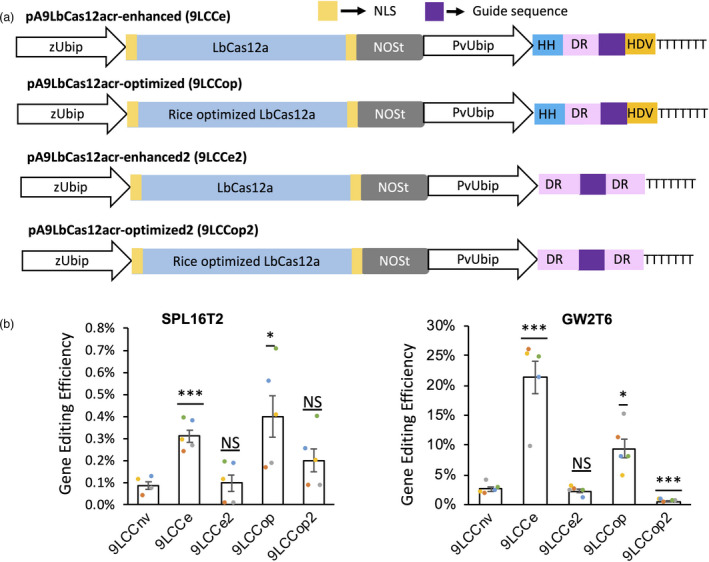
Improving the gene editing efficiency of LbCas12a. (a) Schematic illustration of the modified LbCas12a constructs. Compared to 9LCCnv, in all constructs, the wheat U6 promoter was replaced by the PvUbip promoter. 9LCCe and 9LCCop have hammerhead (HH) and hepatitis delta virus (HDV) ribozymes flanking the 5′ and 3′ ends of crRNA to facilitate its processing; 9Lcce2 and 9LCCop2 have one extra direct repeat (DR) after the 3′ end of the guide sequence; 9LCCe and 9LCCe2 have the human codon‐optimized LbCas12a whilst 9LCCop and 9LCCop2 have the plant codon‐optimized LbCas12a. (b) The comparison of gene editing efficiency amongst the LbCas12a‐based constructs. The performance of these constructs was calculated for the SPL16T2 and GW2T6 targets, which are conserved in all three wheat genomes, using data from four or five biological replicates. The mean ± standard error for each construct is shown on the graphs with the result of each replicate depicted as coloured dots on the bar plots. The gene editing efficiency was normalized by the protoplast transformation efficiency. Student’s t test was applied to assess the significance of differences in gene editing efficiency between 9LCCnv and 9LCCe, 9LCCe2, 9LCCop or 9LCCop2; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; NS P > 0.05.
