Figure 6.
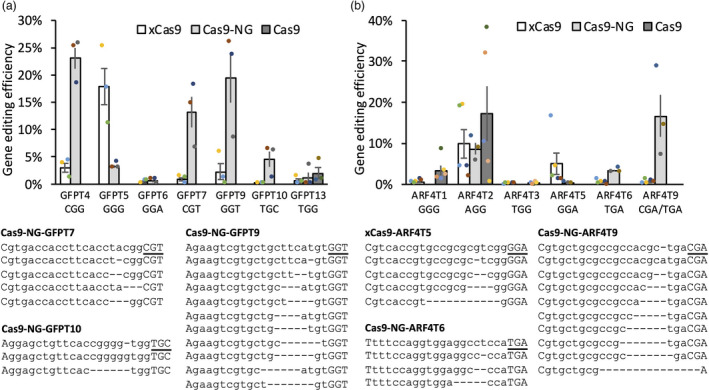
Mutations induced by the Cas9 variants in the wheat genome. (a) Comparison of the xCas9 and Cas9‐NG editing efficiency for targets located within the GFP gene. The calculation of gene editing efficiency are based on three biological replicates. (b) Comparison of the xCas9 and Cas9‐NG editing efficiency for targets located within the TaARF4 genes. Because target sites were conserved across all three wheat genomes, the assessment of gene editing efficiency are based on the mapped reads pooled from all three wheat genomes. The gene editing efficiency was normalized by the protoplast transforming efficiency. The results were based on three to five biological replicates. The data in both (a) and (b) are shown as mean ± standard error, the result from each replicate is shown as coloured dots on the bar plots. The representative mutated reads for the Cas9‐NG’s targets GFPT7, GFPT9, GFPT10, ARF4T6, and ARF4T9, and the xCas9’s target ARFRT5 are shown under the bar plots. The first row for each target is wild type. The PAM sequence is underlined. The target sequences are shown in lower‐case letters. The deleted nucleotides are shown as ‘‐’.
