Figure 1.
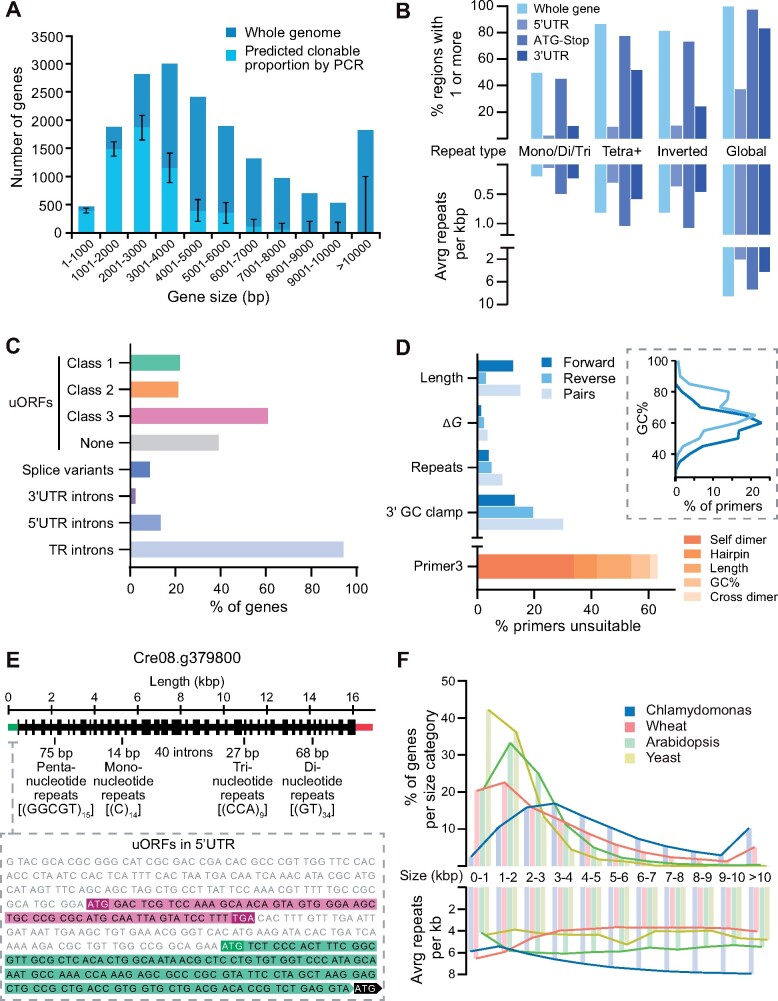
Chlamydomonas nuclear genes are often large, complex, or misannotated, affecting PCR-based cloning attempts and transgene expression success. A, The distribution of gene sizes for the 17,741 genes in the Chlamydomonas nuclear genome (dark blue). Gene sizes are measured from the start of the 5′-UTR to the end of the 3′-UTR. Within each size category, the predicted proportion amenable to PCR-based cloning is shown in light blue. These proportions were extrapolated from cloning success for 624 CCM-related genes from Mackinder et�al. (2017) in which PCR-based cloning was used to amplify the ATG-Stop region of each gene, excluding any UTRs. The strong size-dependence of ATG-Stop cloning efficiency seen in 2017 indicates that 68% of the genome would be challenging to clone. 95% confidence intervals for the predicted clonable proportions of each size category were calculated using the Wilson score interval method. No genes over 8,000 bp are predicted to be clonable by PCR although only a handful of regions of these sizes were tested in 2017 giving rise to the large confidence intervals for these categories. B, Genome-wide sequence complexity, as indicated by the presence of one or more repetitive sequences and frequency of repeats per kbp in each gene (pale blue). Values are also given for repeats localized to the 5′-UTR (light indigo), ATG-Stop (indigo), and 3′-UTR (dark indigo) within each gene. Note that while all 17,741 genes contain a start-to-stop region, not all genes contain a 5′-UTR and/or 3′-UTR, so the percentages presented for these are relative to totals of 17,721 and 17,717, respectively. Simple repeats are shown in the left three categories. Mono/di/tri refers to tandem repeats with a period length of one, two or three; tetra+ refers to all oligonucleotide tandem repeats with a period length of 4 or more and a total length ≥20 bp. Combining whole-gene counts for mono-, di-, tri-, and tetra+ produces an average value of 1.07 tandem repeats per kbp. Inverted repeats refer to short (20–210 bp) sequences that have the potential to form secondary structures by self-complementary base pairing. About 836 genes were free from detectable tandem and inverted repeats under our criteria, most of which are small, with an average length of 1,766 bp. Global repeats refer to repetitive sequences masked by the National Centre for Biotechnology Information (NCBI) WindowMasker program (Morgulis et�al., 2006), which includes both longer, non-adjacent sequences and shorter, simple repeats (see Methods section). All genes contained detectable repetitive regions using the default WindowMasker settings, with an average of 40.07 per gene. UTR data are based on gene models from Phytozome (version 5.5). C, Gene features that complicate correct transgene expression. Top four bars illustrate potential misannotation of functional start sites in the genome shown by the percentage of genes containing one or more uORFs of each class (see text). Note that some genes contain multiple classes of uORF. Shown below this is the percentage of Chlamydomonas genes with multiple transcript models (splice variants), and those containing introns in the UTRs and TRs (between start and stop codons). uORF data are from Cross (2015). Splice variant and intron data are based on gene models from Phytozome (version 5.5). D, Analysis of a set of ATG-Stop PCR primers designed to clone every gene in the genome from start to stop codon using gDNA as the template (Mackinder et�al., 2017). Many primers are predicted to be unsuitable for efficient PCR, as shown by the percentage of forward (dark blue) and reverse (light blue) primers that breach various recommended thresholds associated with good primer design. Pairs (pale blue) are shown for which one or both primers breach the respective thresholds. Thresholds shown pertain to length, secondary structure stability, tandem repeats, and 3′-GC content. The inset shows the distribution of GC content of primers in the dataset, illustrating a clear trend in higher GC content at the 3′-end of coding sequences. Below this, the given reason for rejection of primers by the Primer3 check_primers module is shown in orange. Dimer and hairpin values refer to primers rejected for “high end complementarity” and “high any complementarity” errors, respectively. E, Annotated gene structure of Cre08.g379800. The gene encodes a predicted protein of unknown function but shows examples of several sequence features that contribute to sequence complexity. The unspliced sequence is 16,892 bases long with a GC content of 64.3%. The 41 exons are shown as regions of increased thickness, with 40 introns between them, the annotated 5′-UTR in green (left) and the 3′-UTR in red (right). Labels denote selected examples of simple repeats throughout the gene. The inset shows the 5′-UTR sequence, displaying examples of two classes of uORFs (see text); Class 3 is highlighted in magenta and Class 1 in green. For simplicity, only one of the seven class 3 uORFs are shown in full. Cre08.g379800 was successfully cloned and tagged using recombineering. F, A comparison of gene size and complexity between Chlamydomonas, bread wheat (Triticum aestivum), A. thaliana and Saccharomyces cerevisiae. Gene sizes were binned as in (A), and the average number of global repeats kbp masked by the NCBI WindowMasker program was counted for genes in each size category (Morgulis et�al., 2006). Genes were measured from the start of the 5′-UTR to the end of the 3′-UTR.