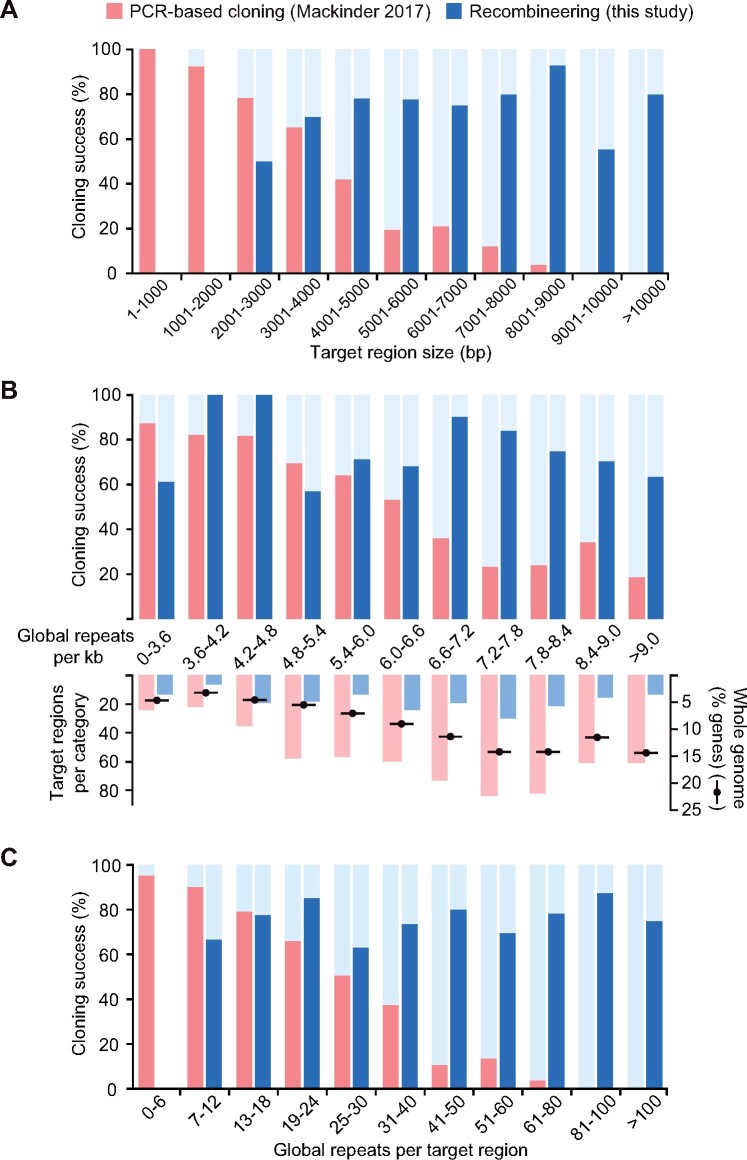
Figure 3.
Our recombineering pipeline is target gene size independent and tolerant of sequence complexity. A, The size distribution of successfully PCR-cloned coding sequences (Mackinder et�al., 2017; red) or recombineered regions (this study; blue) are shown. Regions cloned by recombineering include ∼2 kbp of flanking DNA upstream of the annotated start codon to incorporate native 5′-promoter sequences. A severe drop in PCR-based cloning efficiency can be seen for templates >3-kbp long, whereas recombineering cloning efficiency does not show size dependency. No recombineering target regions were less than 2,000-bp long due to inclusion of native 5′-promoter sequences. B, As above but showing the dependence of cloning success on the per-kilobase frequency of repeats masked by the NCBI WindowMasker program with default settings (Morgulis et�al., 2006). The number of target regions per repeat category is shown beneath this, overlaid with the percentage of Chlamydomonas genes in each category. The distribution of targets for this study and our previous PCR-based cloning attempt (Mackinder et�al., 2017) gives a reasonably close representation of the whole-genome distribution. Almost a third of nuclear genes contain 7.2–8.4 repeats per kbp; this peak corresponds to a clear drop in PCR-based cloning efficiency, but to a high recombineering efficiency of 75–85%. Data for repeats per kbp were continuous and there are no values present in more than one category. C, As above but showing the number of simple and global repeats masked by WindowMasker per template. Data are binned to provide a higher resolution for the lower value categories, since the targets for PCR-based cloning were enriched in targets with low numbers of repeats. As in (A), a severe negative trend in PCR-based cloning efficiency can be seen, reflecting a strong positive correlation between repeat number and region size. No negative association is present for recombineering cloning efficiency, likely illustrating the benefit of avoiding size- and complexity-associated polymerase limitations. No recombineering target regions contained fewer than six repeats.