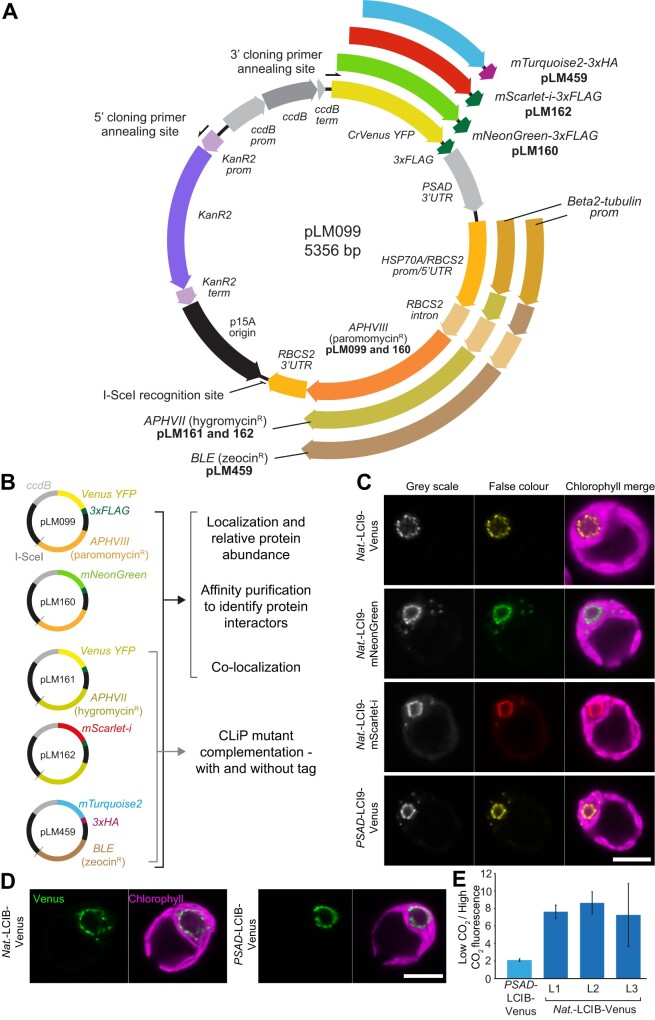
Figure 5.
Development and application of different recombineering vectors to obtain novel biological insights into Chlamydomonas biology. A, Plasmid map for pLM099 and derivative recombineering vectors. PCR amplification with 5′- and 3′-cloning primers at the annealing sites shown results in a ∼4.6-kbp linear cassette for recombineering target genes in-frame with a fluorescent protein and affinity tag. For each recombineering vector, the fluorescent protein sequence is preceded by a flexible linker (GGLGGSGGR) and followed by a tri-glycine linker prior to the affinity tag. The PSAD 3′-UTR terminates all four fluorescent protein-affinity tag cassettes. The RBCS2 3′-UTR terminates all three Chlamydomonas selection cassettes. The same RBCS2 intron is present in all three Chlamydomonas selection cassettes but is only inter-exonic in the hygromycin and zeocin-resistance cassettes. B Additional vectors for tagging with different fluorophores and for complementation of Chlamydomonas library mutants generated using insertion of the AphVIII paromomycin resistant gene. C, Localization of LCI9 with different fluorescence protein tags. LCI9 was recombineered with its native promoter (Nat.) using pLM099, pLM160, and pLM162. A previously developed line cloned by PCR and using the constitutively expressed PSAD promoter is shown for comparison (PSAD-LCI9-Venus). Scale bar: 5 �m. D, A comparison of the low CO2 upregulated gene LCIB cloned with its native promoter via recombineering versus LCIB under the control of the constitutive PSAD promoter. Cells were grown and imaged at atmospheric CO2 levels. Scale bar: 5 �m. E, Relative change in LCIB-Venus fluorescence between high (3% v/v) and low (0.04% v/v) CO2 when expressed from the constitutive PSAD promoter versus expression from the native LCIB promoter. Data are shown for three independent native LCIB promoter lines (L1–L3). Error bars are standard error of the mean.