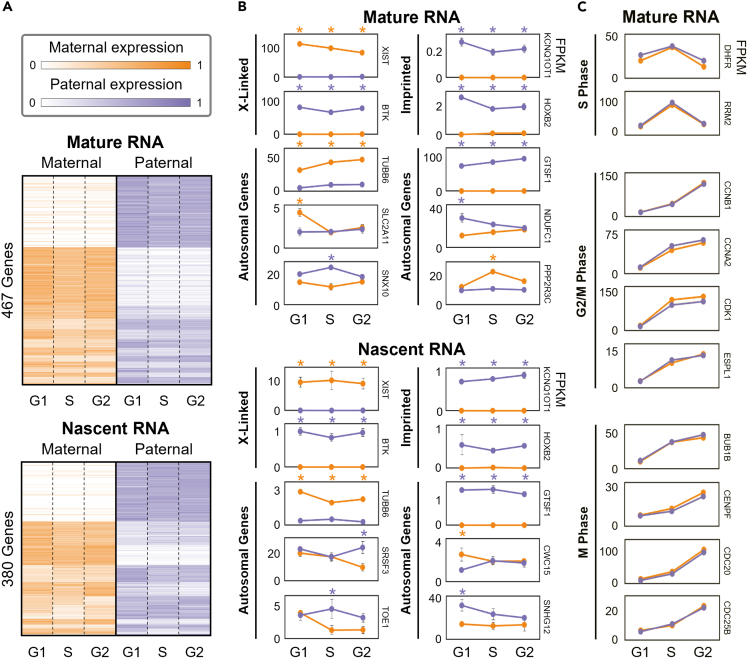
Figure 3.
Allele-specific mature and nascent RNA expression
(A) Differentially expressed genes’ maternal and paternal RNA expression through the cell cycle. Expression heatmaps are average FPKM values over three replicates after row normalization. Genes are grouped by their differential expression patterns (Figure S1).
(B) Representative examples of X-linked, imprinted, and other autosomal genes with allelic bias. Top and bottom sections of (A) and (B) show mature RNA levels (RNA-seq) and nascent RNA expression (Bru-seq), respectively.
(C) Examples of cell cycle regulatory genes’ mature RNA levels through the cell cycle. These genes are grouped by their function in relation to the cell cycle and all exhibit CBE but none have ABE. All example genes in (B) and (C) reflect average FPKM values over three replicates, and ABE in a particular cell cycle phase is marked with an orange or purple asterisk for maternal or paternal bias, respectively. Within this figure, G2 includes both G2 and M phase. Any plots with no visible error bars in (B) and (C) have errors smaller than the marker at each cell cycle phase.