Figure 4.
Haplotype phasing of Hi-C data
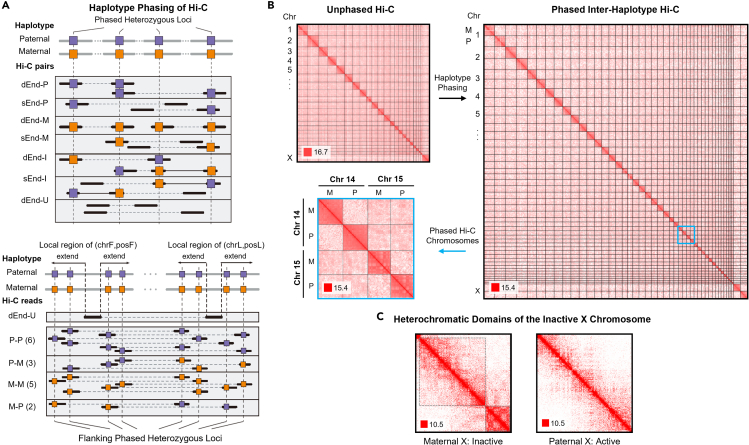
(A) HaploHiC separates paired-end reads into groups based on parental origin determined through SNVs/InDels (left, method details). Reads are grouped by: (i) reads with one (sEnd-P/M) or both ends (dEnd-P/M) mapped to a single parent, (ii) reads are inter-haplotype, with ends mapped to both parents (d/sEnd-I), and (iii) reads with neither end mapped to a specific parent (dEnd-U). Abbreviations within this figure are defined as follows: dEnd, double-end; sEnd, single-end; U, unmapped; P, paternal; M, maternal; I, inter-haplotype; chrF/posF, mapped chromosome and position of forward end after sorted; chrL/posL, mapped chromosome and position of latter end after sorted. An example of a paired-end read (dEnd-U) with no SNVs/InDels has its origin imputed using nearby reads (right, method details). A ratio of paternally and maternally mapped reads is found in a dynamically sized flanking region around the haplotype-unknown read’s location (method details). The ratio then determines the likelihood of the haplotype-unknown read’s origin. This example visualization shows a slight bias toward Hi-C reads with a paternal origin, but the majority of our Hi-C data has stronger biases than what is shown here.
(B) Whole-genome Hi-C of GM12878 cells (top left). Inter-haplotype and intra-haplotype chromatin contacts after phasing Hi-C data using HaploHiC (right). Chromosomes 14 and 15 highlight inter- and intra-chromosome contacts within and between genomes (bottom left). Visualized in log2 scale 1 Mb resolution in G1.
(C) Haplotype phasing illustrates that the inactive maternal Chromosome X is partitioned into large heterochromatic domains, outlined in dotted black boxes. Visualized in log2 scale 100 kb resolution in G1.