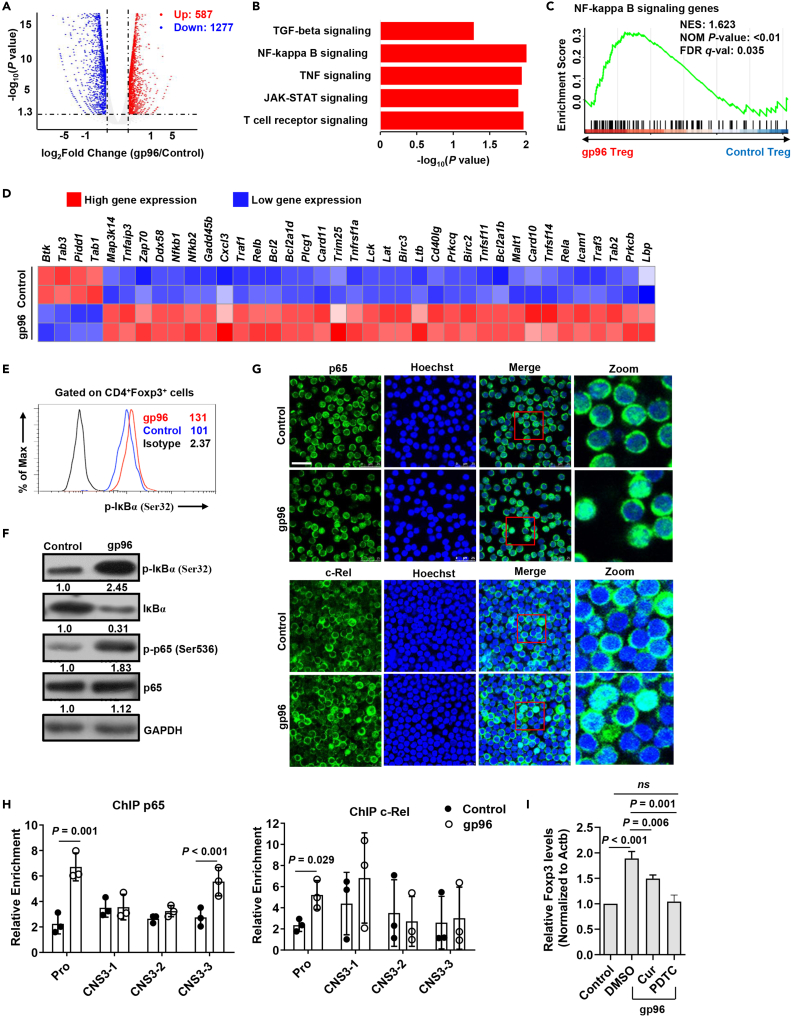
Figure 5.
Gp96 induces Foxp3 expression via NF-кB pathway
(A) Volcano map depicting genes upregulated (red) or downregulated (blue) 2-fold or more in Treg cells.
(B) Gene Ontology (GO) terms of the differentially expressed genes in gp96-treated Tregs compared with control Tregs.
(C) Gene set enrichment analysis (GSEA) of NF-кB signaling gene set in gp96-treated Tregs relative to expression in control Treg cells.
(D) Heatmap of NF-кB signaling pathways (Log2 fold change) that are differentially expressed.
(E) FACS analysis of IкB phosphorylation in Foxp3+ Tregs treated with 100 μg/mL of gp96 or saline as control in vitro for 0.5 h.
(F) Western blot analysis of IкB-α and p65 phosphorylation in sorted Tregs treated as in (E).
(G) p65 and c-Rel were detected using immunofluorescent staining. Scale bar, 25 μm.
(H) Chromatin immunoprecipitation (ChIP) analysis of p65 and c-Rel binding at Foxp3 promoter region. Chromatin obtained from spleen Tregs (1×106) of gp96-immunized or control mice were immunoprecipitated with anti-p65 and anti-c-Rel antibodies, followed by real-time PCR analysis.
(I) mRNA levels of Foxp3 in Tregs treated with 100 μg/mL gp96 in vitro and curcumenol or PDTC were analyzed using real-time PCR after 24 h. The data are representative of two independent experiments with similar results. n = 5 mice/group. Mean ± SD is shown. The Student's t-test was used for statistical analysis. P < 0.05 was considered statistically significant. ns = not significant.