Figure 5. Minimal 46‐aa region is required for Nup50 NPC localization.
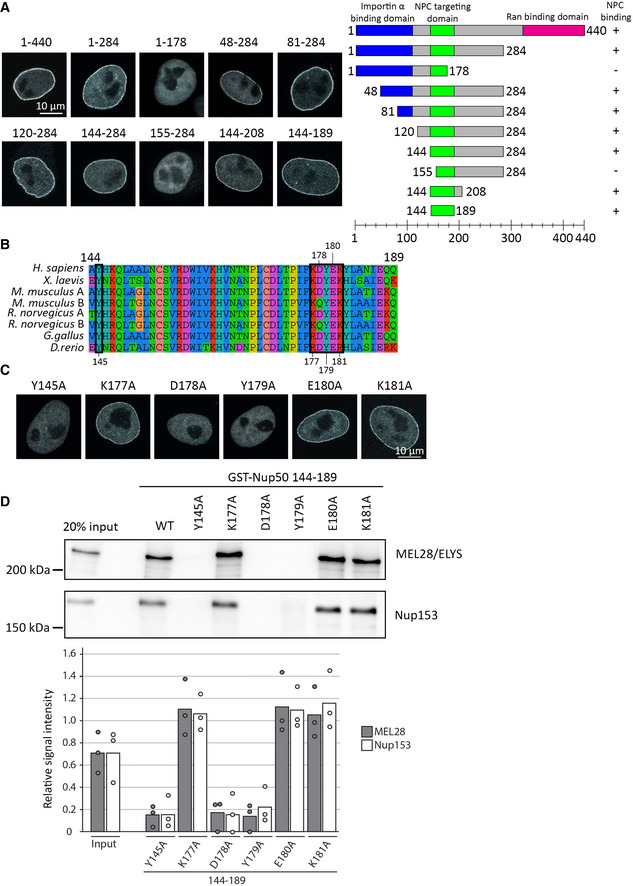
- HeLa cells were transfected with EGFP‐Nup50 and different truncations. After 24 h, cells were shortly pretreated with 0.1% Triton X‐100 in PBS, fixed with 4% PFA, and analyzed by confocal microscopy (left panel). Scale bar: 10 µm. The graph (right panel) shows the Nup50 domains (blue: importin α interaction domain, green: NPC‐targeting domain, and pink: Ran‐binding domain), and the truncation tested. Ability to bind NPCs is indicated on the right.
- Sequence alignment of Nup50 144–189, species are indicated on the left, the black boxes highlight residues tested by single‐point mutation in (C) and (D), and the residue number are shown above or below the alignment. The color scheme indicates the type of amino acids according to the alignment software default setting.
- HeLa cells were transfected with EGFP‐Nup50 comprising different point mutations in the minimal NPC‐binding region and analyzed as in (A). Scale bar: 10 µm.
- GST fusion constructs of the Xenopus Nup50 minimal NPC binding fragment (aa 144–189) comprising no or single‐point mutations were incubated with Xenopus egg extracts. Starting material as well as bound proteins were analyzed by Western blotting with indicated antibodies. The quantitation shows the average MEL28/ELYS and Nup153 bead bound signal from three independent experiments, normalized to the signal of their respective wild‐type GST fusion (see Appendix Fig S1D for a gel showing the GST baits). Data points from individual experiments are indicated.
Source data are available online for this figure.
