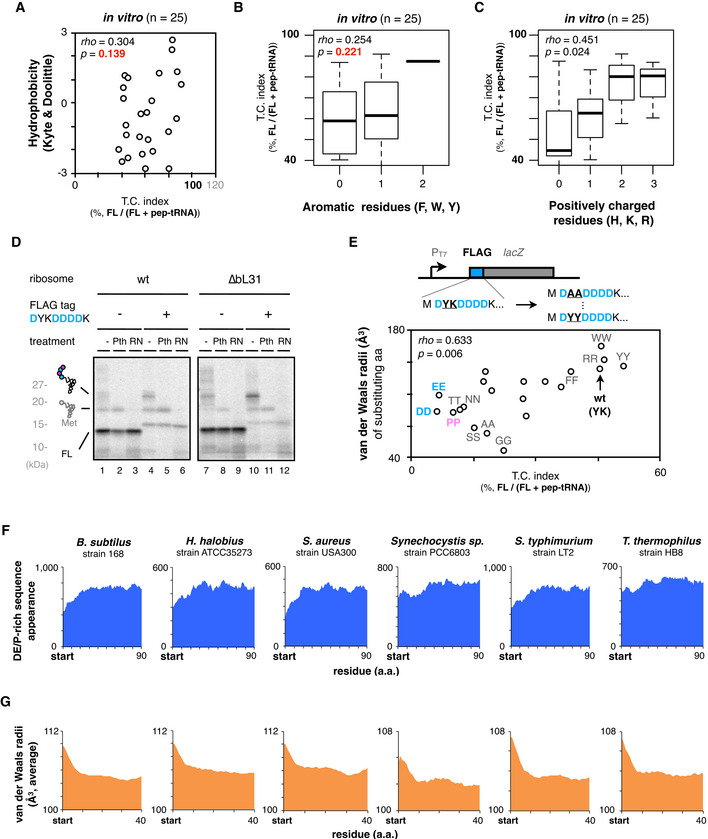
Figure EV2. IRD‐counteracting bulky amino acids near the PTC and genome‐wide biases in amino acids distributions.
-
ATwo‐dimensional plots of downstream translation continuation of the 5 aa‐EPDP constructs (TC index) and the average hydrophobicity of inserted amino acid sequences. The result of Spearman's correlation test is also shown.
-
B, CBoxplot of the TC index and the appearance of aromatic (B) or positively charged residues (C) of inserted amino acid sequences. The result of Spearman's correlation test is also shown.
-
DTranslation of the N‐terminal FLAG tag‐lacZ construct induces the accumulation of abortive peptidyl‐tRNAs. In vitro translation was performed using standard (lane 1–6) or a customized PURE system equipped with ∆bL31 ribosome (lane 7–12) in the presence of 35S‐methionine, and polypeptide products were separated by neutral pH SDS–PAGE.
-
EDependency of the molecular radii of amino acids on the translation continuation using FLAG tag as a model IRD‐prone sequence. Second tyrosine and third lysine residues of the N‐terminal FLAG tag‐lacZ were replaced with 20 kinds of dipeptide motif as described in the upper schematic. FLAG tag variants were translated in vitro, and the correlation between the downstream translation continuation and the van der Waals molecular radii of substituted amino acids are plotted in two‐dimensional. The result of Spearman's correlation test is also shown.
-
F, GAppearance of the DE/P‐rich sequences (F) or average van der Waals molecular radii of 5‐aa moving window (G) at the N‐terminal regions in the total ORFs of various bacterial species.
