Figure 4. Effect of mutations in the constriction site within the exit tunnel on the IRD counteraction.
-
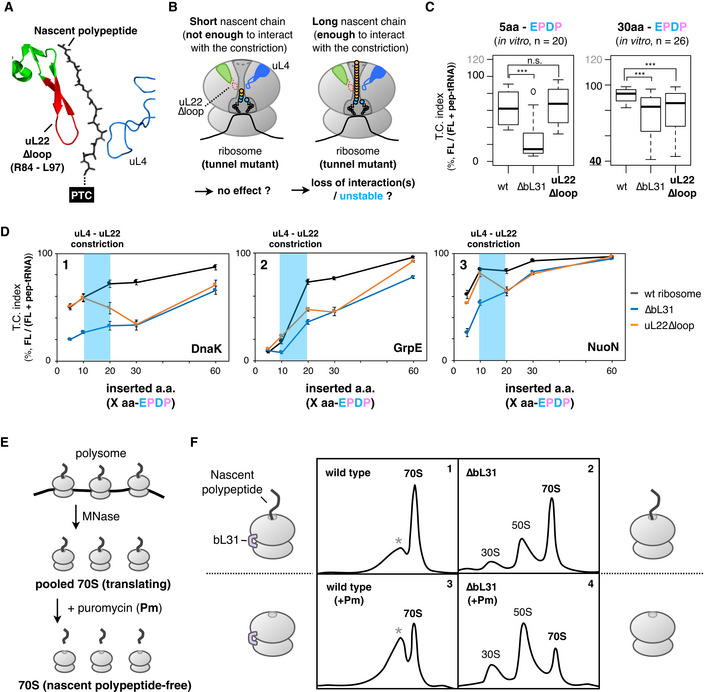
AStructure of the uL4‐uL22 constriction site (PDB: 4V5H). Deleted residues in the uL22∆loop mutation are indicated in red.
-
BSchematic of the working hypothesis that stabilization of the translating ribosome depends on interactions between the ribosomal tunnel and the tunnel‐occupying nascent polypeptide.
-
CInserted amino acid length‐dependent IRD‐counteracting effects in the uL22∆loop‐ribosome. Messenger RNAs encoding a 5‐aa (left) or 30‐aa (right) insertion and EPDP (used in Fig 2) were individually translated by customized PUREfrex including the indicated ribosome variants. Evaluated downstream translation continuation indices of wild‐type, ∆bL31, or uL22∆loop ribosomes are represented by boxplots using R software. Box portion and central band are described according to the 25th percentile and the median, respectively. ***P‐value < 0.001, n.s.: no significant difference (Welch’s t test).
-
DPreceding nascent polypeptide‐length dependency on the IRD counteraction in the ribosome variants. The N‐terminal regions of E. coli DnaK (panel 1), GrpE (panel 2) or NuoN (panel 3) were utilized as preceding nascent polypeptides. The mean values ±SE estimated from three independent technical replicates are shown.
-
EExperimental design to evaluate the stability of the translating ribosomes. MNase: micrococcal nuclease.
-
FStability of the 70S ribosome complex with or without a tunnel‐occupying nascent polypeptide in wild‐type (panels 1 and 3) or ∆bL31 (panels 2 and 4) cells. Pooled 70S translating ribosomes with (panels 3 and 4) or without (panels 1 and 2) the puromycin treatment were fractionated by SDG in the presence of 10 mM magnesium ion. Recorded A254 values are shown. The peaks indicated as the asterisk (*) in panels 1 and 3 seem to be an aberrant form of the 70S ribosome due to the MNase treatment.
