Figure 8.
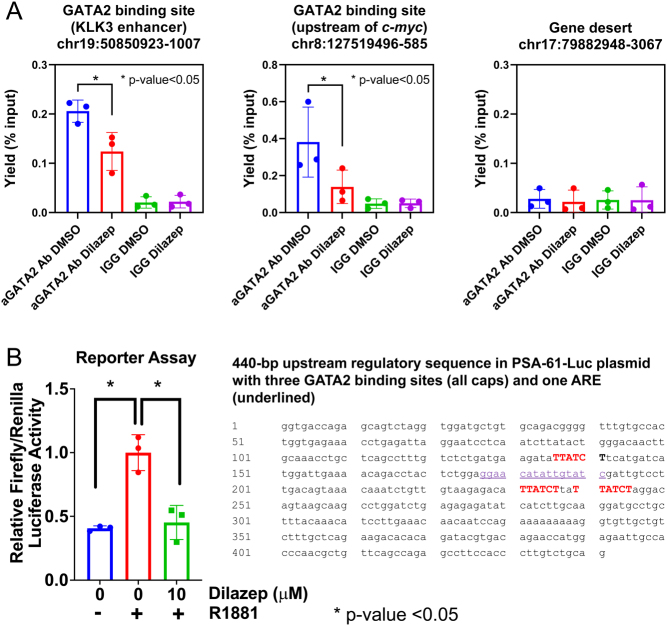
(A) Dilazep inhibits GATA2 recruitment to chromatin. LNCaP cells were treated with 10 µM of dilazep for 16 h and subjected to chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) analysis for the presence of GATA2 on chromatin. Chromatin was fixed with formaldehyde, then DNA was extracted and sonicated into short fragments. GATA2-associated DNA was immunoprecipitated using anti-GATA2 or rabbit IgG as a negative control and analyzed by quantitative PCR as per Materials and methods. We selected two GATA2 binding sites based on prior GATA2 ChIP-seq profiles. The genomic coordinates for these sites are as indicated. Both sites harbor GATA2 binding sequences. Error bars indicate standard deviations for triplicates. (B) Dilazep suppresses transcriptional activity in a reporter assay based on the KLK3/PSA gene promoter/enhancer. 22Rv1 cells were transfected with reporter vector (pGL3/PSA61-Luc) and vectors encoding for Renilla luciferase (pGL4.75, Promega) and GATA2 (pcDNA3-GATA2) using jet PRIME transfection kit (Polyplus) according to the manufacturer’s protocol. The pGL3/PSA61-Luc harbors several GATA2 binding sites (those within a 440-bp regulatory sequence are shown here; for more details, please see Supplementary Text file) and three AREs (one of which is highlighted; two additional AREs are located right before the luciferase gene start site, please see Supplementary Text file). After 24 h, cells were treated with 5nM R1881 and either 10 µM dilazep or DMSO for 24 h. Luciferase activity in cells was measured with Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer protocol. Relative firefly-to-Renilla luciferase activity from three independent experiments was plotted as the means ± s.d. All bar plots were generated using GraphPad Prism 8. A full color version of this figure is available at https://doi.org/10.1530/ERC-21-0085.

 This work is licensed under a
This work is licensed under a