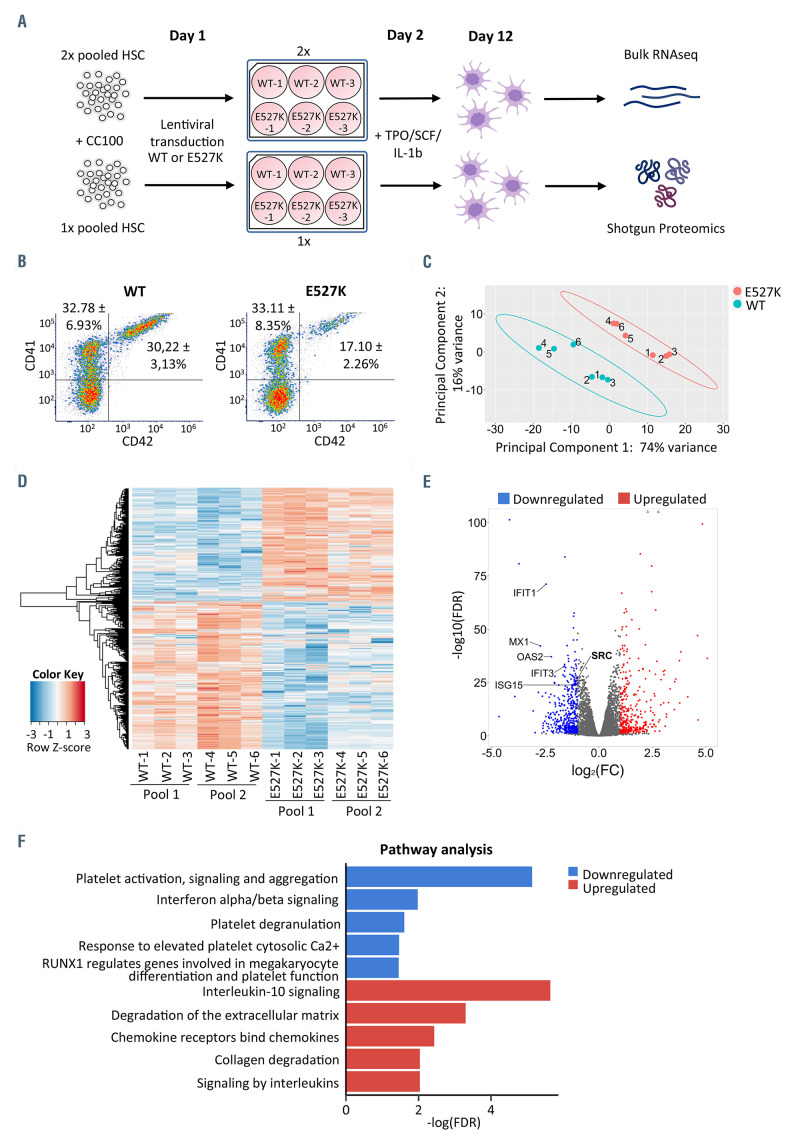
Figure 1.
Bulk RNA sequencing of WT-SRC and E527K-SRC megakaryocytes. (A) A total of three hematopoietic stem cell (HSC) pools were isolated from peripheral blood from 15 healthy controls and cultured in HSC amplification medium (with cytokine cocktail 100). On day 1 after isolation, HSC were transduced in triplicate with either wild-type SRC (WT-SRC) or E527K-SRC lentiviral vectors. On day 2, differentiation to megakaryocytes (MK) was initiated upon addition of the cytokines TPO, SCF and IL1b. Day 12 mature MK were collected for bulk RNA sequencing and shotgun proteomics. (B) Flow cytometry of day 12 MK stained for CD41 (forward scatter) and CD42 (side scatter). The % of CD41+ and CD41/CD42+ MK are shown for WT-SRC and E527K-SRC cells as mean and standard deviation for 3 cultures. (C) Principal component analysis of 6 WT-SRC and 6 E527K-SRC MK samples used for RNA sequencing. (D) Heatmap visualizing the 852 differentially expressed genes (DEG) identified in E527K-SRC vs. WT-SRC MK samples (red, upregulated genes; blue, downregulated genes). (E) Volcano plot showing the 852 DEG detected in E527K-SRC MK organized according to log2(fold-change [FC]) and -log10(false discovery rate [FDR]). (red, upregulated genes with FDR<0.05 and log2FC>1; blue, downregulated genes with FDR<0.05 and log2FC<-1). (F) The 852 DEG were used for Reactome pathway analysis. Top 5 downregulated (blue) and upregulated (red) pathways are shown as arranged by their -log(FDR) value.