Abstract
Background
Gliomas are primary aggressive brain tumors with poor prognoses. Oxidative stress plays a crucial role in the tumorigenesis and drug resistance of gliomas. The aim of the present study was to use integrated bioinformatics analyses to evaluate the prognostic value of oxidative stress-related genes (OSRGs) in glioma.
Material/Methods
Disease- and prognosis-associated OSRGs were identified using microarray and clinical data from the Chinese Glioma Genome Atlas database. Functional enrichment, gene-gene interaction, protein-protein interaction, and survival analyses were performed in screened OSRGs. The protein expression was validated by the Human Protein Atlas database. A risk score model was constructed and verified through Cox regression, receiver operating characteristic curve, principal component, and stratified analyses. The Cancer Genome Atlas (TCGA) database was used for external validation. A nomogram was constructed to facilitate the clinical application.
Results
Twenty-one disease-associated and 14 prognosis-associated OSRGs were identified. Enrichment analyses indicated that these signature OSRGs were involved in tumorigenesis and drug resistance of glioma. The risk score model demonstrated a significant difference in overall survival between the high- and low-risk groups. The area under the curve and hazard ratio (1.296) revealed the independent prognostic value of the model. The model exhibited good predictive efficacy in the TCGA cohort. A clinical nomogram was constructed to calculate survival rates in glioma patients at 1, 3, and 5 years.
Conclusions
Our comprehensive study indicated that OSRGs were valuable for prognosis prediction in glioma, which provides a novel insight into the relationship between oxidative stress and glioma and a potential therapeutic strategy for glioma patients.
Keywords: Computational Biology; Glioma; Models, Statistical; Oxidative Stress; Prognosis
Background
Gliomas are one of the most common malignant primary brain tumors in adults [1,2]. According to the 2016 World Health Organization (WHO) Classification [3], they are classified into 4 grades (I, II, III, and IV), based on histopathologic appearance and specific molecular parameters including IDH mutation [4], 1p/19q codeletion [5], and MGMT promoter methylation [6]. Distinguishing different grades and types of gliomas has clinical significance for identifying an effective treatment strategy and assessing prognosis. Generally, patients with low-grade gliomas (grades I and II) have a relatively favorable prognosis after surgery. In contrast, in patients with grade IV tumors (glioblastoma multiforme [GBM]), the prognosis remains very poor even with comprehensive and aggressive therapy, and the median overall survival (OS) is no more than 2 years [1]. Given the heterogeneous prognosis of this disease, accurate diagnosis and classification are of the utmost importance for optimizing therapy. Therefore, it is important to explore the mechanisms of carcinogenesis and identify better prognostic indicators for glioma patients.
Redox reactions are ubiquitous and fundamental biological processes among cells. The generation of reactive oxygen species (ROS) including superoxide anions, hydrogen peroxide (H2O2), and hydroxyl radicals (•OH) is an unavoidable consequence of metabolism. ROS overproduction can cause DNA damage, protein oxidation, and lipid peroxidation. To avoid these negative effects, glutathione (GSH), thioredoxin (TXN), and NADPH play pivotal roles in countering oxidative stress [7]. Oxidative stress, in which oxidative stress-related genes (OSRGs) participate, is a state of redox disequilibrium and profoundly affects various functions and processes, including cell proliferation, cell differentiation, angiogenesis, and metabolism [8–11]. It has been implicated in the pathophysiology of several disorders, such as Alzheimer disease [12], diabetes mellitus [13], and atherothrombosis [14]. Oxygen and ROS are the critical components of oxidative stress.
The effects of ROS on gliomas are complex. On one hand, they can alter and reconstruct the genetic material, resulting in tumorigenesis. Grades II and III glioma cells demonstrate mitochondrial DNA mutations with higher oxidative stress [15]. On the other hand, excessive ROS is cytotoxic. If glioma cells cannot promptly eliminate excessive ROS, mitochondrial disruption and metabolic disturbance can occur, which induces the death of the glioma cells [7,8,16]. Typically, the oxygen-involved metabolism is vigorous in glioma and GBM to fuel the increased energy expenditure in the generation, proliferation, and differentiation of the cancer cells [17,18]. Oxidative stress is also associated with drug resistance in glioma cells. lncRNA H19 induced by oxidative stress confers temozolomide resistance by activating NF-κB signaling [19].
This suggests that the oxidative stress process and the level of expression of OSRGs in glioma cells may influence the prognosis and survival of glioma patients. Hence, identifying the potential value of OSRGs may help predict clinical outcomes and offer therapeutic strategies for glioma patients.
In the present study, we screened and focused on candidate OSRGs. The Chinese Glioma Genome Atlas (CGGA) database was used to explore the OSRGs relevant to clinical prognosis. We then established a mathematical risk score model of the expression level of these genes to predict prognoses in glioma patients. The reliability of this model was verified through external and internal validation. Further, a clinical nomogram was constructed to facilitate clinical application. It is hoped that our study can provide new insights into understanding the complex biological actions of OSRGs in glioma cells and lead to novel strategies for the treatment of glioma.
Material and Methods
Retrieval of Oxidative Stress-Related Genes
OSRGs were searched from the Gene Set Enrichment Analysis (GSEA) database (https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp). Then, “oxidative_stress” was designated as a keyword and 29 gene sets were found. The “GO_RESPONSE_TO_OXIDATIVE_STRESS” gene set (systematic name: M16361) was chosen to ensure gene data integrity. It consisted of 453 OSRGs of the crucial biological processes that contribute to modification of the activity or state of a cell or organism due to oxidative stress, including cell movement, secretion, enzyme production, and gene expression.
Acquisition and Processing of Gene Expression Profiles and Clinical Data
Gene expression profiles for and corresponding clinical information from glioma patients were downloaded from the CGGA database. The patients were treated at Beijing Tiantan Hospital, Sanbo Hospital in Beijing, Tianjin Medical University General Hospital, The First Affiliated Hospital of Nanjing Medical University, Harbin Medical University, and China Medical University. All patients for whom information existed in the CGGA database were diagnosed with gliomas and their data were reviewed by independent neuropathologists, based on the 2016 World Health Organization classification. Written informed consent was obtained from all patients. The mRNA microarray data from 1018 glioma samples (693 in batch 1 and 325 in batch 2) and 20 normal samples were collected and preprocessed using the “sva” and “limma” R packages to eliminate the batch effect [20,21]. The clinical information consisted of survival data (survival status and OS) and other clinicopathological parameters, such as sex; age; grade; histology; primary, recurrent, or secondary (PRS) tumor type, radiotherapy status, chemotherapy status (temozolomide-treated or -untreated), and IDH mutation, 1p19q codeletion, and MGMT promoter methylation statuses.
Identification and Enrichment Analysis of Disease-Associated OSRGs
The Wilcox signed-rank test was performed to identify OSRGs that were expressed differently in glioma and normal samples. A criterion was set for screening with a false discovery rate (FDR) <0.05 and an |log2 FC| >1 via the “limma” R package [22]. The screened genes were designated as disease-associated OSRGs. The expression levels were illustrated with the “pheatmap” and “ggplot” R packages.
To detect the potential biological and molecular function attributed to these OSRGs, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using “clusterProfiler” R packages [23]. An adjusted P<0.05 was set as the threshold of significance and the enrichment analysis results were presented using “ggplot2” and “GOplot” R packages.
Identification of Prognosis-Associated OSRGs
The survival data and mRNA expression profiles of glioma samples were then combined into a matrix. Univariate and multivariate Cox regression analyses were used to screen differentially expressed OSRGs. The cut-off value was P<0.05. These genes were designated as the prognosis-associated OSRGs.
Exploration of the Relationship of Prognosis-Associated OSRGs
Pearson’s correlation test was used to investigate the interaction associations of prognosis-associated OSRGs. Gene interaction network analyses, including genetic interactions, co-expression, physical interactions, co-localization, and shared protein domains, were performed using GeneMANIA (https://www.genemania.org) [24]. A protein-protein interaction (PPI) network was created using STRING (https://string-db.org/) and Cytoscape software [25,26].
Immunohistochemical Analysis of Prognosis-Associated OSRGs
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/, Version 20.1), an open-access protein expression database containing human transcriptomic and proteomic data, was used to analyze the level of protein expression in OSRGs [27]. Immunohistochemical data were obtained from glioma and glial cells in normal brain tissue to analyze the expression of OSRGs [28].
Risk Score Model Construction and Survival Analysis
The risk score model was generated as the sum (∑) of the coefficient value (Ci) multiplied by the expression value (Ei) of each selected prognosis-associated OSRG.
| (1) |
Based on the median risk score, glioma patients were classified into high- and low-risk groups. To determine the reliability of the model, patients were also divided into multiple subgroups according to various clinicopathological parameters, including grade. IDH mutation, 1p19q codeletion, MGMT promoter methylation, chemotherapy, and radiotherapy statuses. Kaplan-Meier survival analyses were performed to estimate the OS differences using the “survival” and “survminer” R packages.
Independent Prognostic Analysis of the Risk Score Model
To evaluate the independent prediction efficiency of the risk score model, univariate and multivariate Cox regression analyses were carried out and visualized using the “survival” R package. A receiver operating characteristic (ROC) curve analysis was performed and an area under the curve (AUC) score >0.75 represented good model prediction. A principal component analysis (PCA) was conducted with the “limma” and “scatterplot3d” R packages to evaluate the discrimination ability of the model. Stratification analysis was performed to evaluate the relationship between the risk score and other clinical parameters.
External Validation with TCGA Cohort
To assess the external validity of the risk score model, gene expression profiles (FPKM-standardized RNA-seq data) and survival data (survival state and OS) of lower-grade glioma (LGG) as well as GBM were obtained from TCGA GDC Data Portal (https://portal.gdc.cancer.gov/). In total, 668 glioma patients were enrolled for external validation and their risk scores were calculated with the same formulas as previously described. A Kaplan-Meier survival analysis was performed after the samples were classified into 2 risk groups according to their risk scores. Univariate and multivariate Cox regression and ROC analyses were performed to further validate the risk score model in TCGA.
Gene set Enrichment Analysis
To develop a comprehensive understanding of the difference between high- and low-risk groups in the CGGA cohort, GSEA was performed using GSEA software (Version 4.0.1, http://www.broad.mit.edu/gsea/) [29]. Anominal P<0.05 and FDR <0.25 were considered statistically significant.
Nomogram Construction
To facilitate clinical use of results of the analysis, the risk score was combined with other crucial clinical parameters to construct a nomogram using the “rms” R package. The total points indicated the 1-, 3-, and 5-year survival rates for glioma patients
Results
Identification and Functional Annotation of Disease-Associated OSRGs
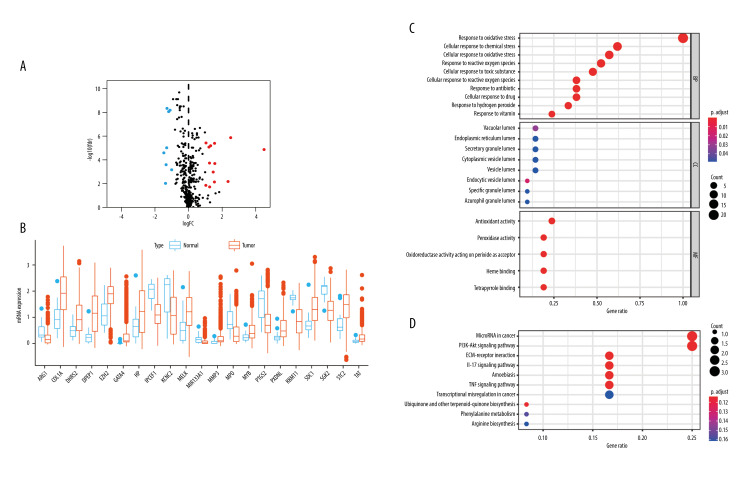
A total of 453 OSRGs were acquired from the “GO_RESPONSE_TO_OXIDATIVE_STRESS” gene set. According to the data on mRNA expression from 1018 glioma samples and 20 normal samples downloaded from the CGGA database, 21 differentially expressed, disease-associated OSRGs were obtained, including 13 upregulated OSRGs (MYB, EZH2, SDC1, MELK, HP, STC2, DHRS2, TAT, COL1A1, PXDNL, MMP3, DPEP1, and GATA4) and 8 downregulated OSRGs (MPO, MIR133A1, ARG1, PTGS2, RBM11, IPCEF1, SGK2, and KCNC2) (Figure 1A, 1B).
Figure 1.
Twenty-one disease-associated, oxidative stress-related genes (OSRGs) and functional enrichment analysis. (A) In a volcano map, red indicates genes that are highly expressed, blue indicates genes with lower expression, and black indicates genes for which there is no significant difference in expression. (B) Differences in gene expression in samples from glioma (red bars) and normal tissue (blue bars). (C) The biological process (BP), cellular components (CC), and molecular functions (MFs) of OSRGs in the Gene Ontology analysis. (D) The potential pathways of OSRGs in the Kyoto Encyclopedia of Genes and Genomes analysis. (R Studio, Version 1.2.5042, RStudio, Inc.).
GO and KEGG functional enrichment analyses were performed with these disease-associated OSRGs. The GO enrichment analysis was divided into 3 sections. In biological processes (BP), these OSRGs were mainly enriched in cellular response to oxidative stress, chemical stress, ROS, toxic substance, hydrogen peroxide, antibiotics, drugs, and vitamins. In cellular components (CC), the OSRGs were primarily involved in the lumens of the endocytic vesicle, vacuole, specific granule, endoplasmic reticulum, azurophil granule, secretory granule, cytoplasmic vesicle, and vesicle lumen. In molecular functions (MFs), the OSRGs were mainly associated with antioxidant, peroxidase, and oxidoreductase; acting on peroxide as an acceptor; and binding of heme and tetrapyrrole (Figure 1C).
In KEGG enrichment analysis, the OSRGs were mainly enriched in ECM-receptor interaction, the interleukin (IL)-17 signaling pathway, microRNAs in cancer, amoebiasis, the tumor necrosis factor (TNF) signaling pathway, the PI3K−Akt signaling pathways, biosynthesis of ubiquinone and other terpenoid−quinones, phenylalanine metabolism, arginine biosynthesis, and transcriptional misregulation in cancer (Figure 1D).
These enrichment analyses revealed that the effect of OSRGs on the tumorigenesis of glioma cells is complex.
Identification of Prognosis-Associated OSRGs
A differentially expressed, disease-associated gene might not necessarily affect the clinical outcome of patients. To explore the relationship between the expression of the differentially expressed OSRGs and prognosis of glioma, 21 screened OSRGs were further filtered. First, a univariate Cox regression analysis was performed and 18 OSRGs were correlated with the prognosis of glioma patients (Table 1). After multivariate Cox regression analysis, 14 prognosis-associated OSRGs were identified, including ARG1, COL1A1, DHRS2, DPEP1, EZH2, GATA4, IPCEF1, KCNC2, MELK, MMP3, PTGS2, PXDNL, RBM11, and TAT (Table 1).
Table 1.
Identification of prognosis-associated OSRGs in glioma samples.
| Gene | Univariate Cox regression | Multivariate Cox regression | Coefficient value | ||
|---|---|---|---|---|---|
| Hazard ratio | P value | Hazard ratio | P value | ||
| ARG1 | 1.765611 | 0.000486 | 1.331385 | 0.09786 | 0.286219 |
| COL1A1 | 2.152233 | 5.99E-47 | 1.283431 | 0.003625 | 0.249537 |
| DHRS2 | 0.60862 | 1.07E-13 | 0.68651 | 4.44E-07 | −0.37613 |
| DPEP1 | 1.702555 | 1.41E-24 | 1.172631 | 0.019606 | 0.15925 |
| EZH2 | 2.787418 | 1.48E-25 | 0.680391 | 0.029804 | −0.38509 |
| GATA4 | 1.938937 | 1.23E-17 | 1.447765 | 4.01E-05 | 0.370021 |
| HP | 1.207974 | 1.83E-05 | – | – | – |
| IPCEF1 | 0.635518 | 1.18E-08 | 1.389578 | 0.007091 | 0.329 |
| KCNC2 | 0.534093 | 1.69E-30 | 0.760656 | 0.000178 | −0.27357 |
| MELK | 2.883717 | 4.28E-58 | 2.964994 | 1.01E-14 | 1.086875 |
| MIR133A1 | 0.400577 | 0.041315 | – | – | – |
| MMP3 | 1.356823 | 0.003276 | 1.250917 | 0.069461 | 0.223877 |
| MYB | 2.261163 | 7.28E-18 | – | – | – |
| PTGS2 | 1.368135 | 6.04E-06 | 0.854968 | 0.096688 | −0.15669 |
| PXDNL | 2.388241 | 9.55E-25 | 0.84379 | 0.148546 | −0.16985 |
| RBM11 | 0.703728 | 1.65E-06 | 1.182832 | 0.061561 | 0.167912 |
| SDC1 | 2.618513 | 7.24E-48 | – | – | – |
| TAT | 0.516778 | 0.000167 | 0.56148 | 0.001091 | −0.57718 |
Eighteen oxidative stress-related genes (OSRGs) were screened using univariate Cox regression analysis. Fourteen OSRGs were screened using multivariate Cox regression analysis.
Investigation of Interactions Among Prognosis-Associated OSRGs
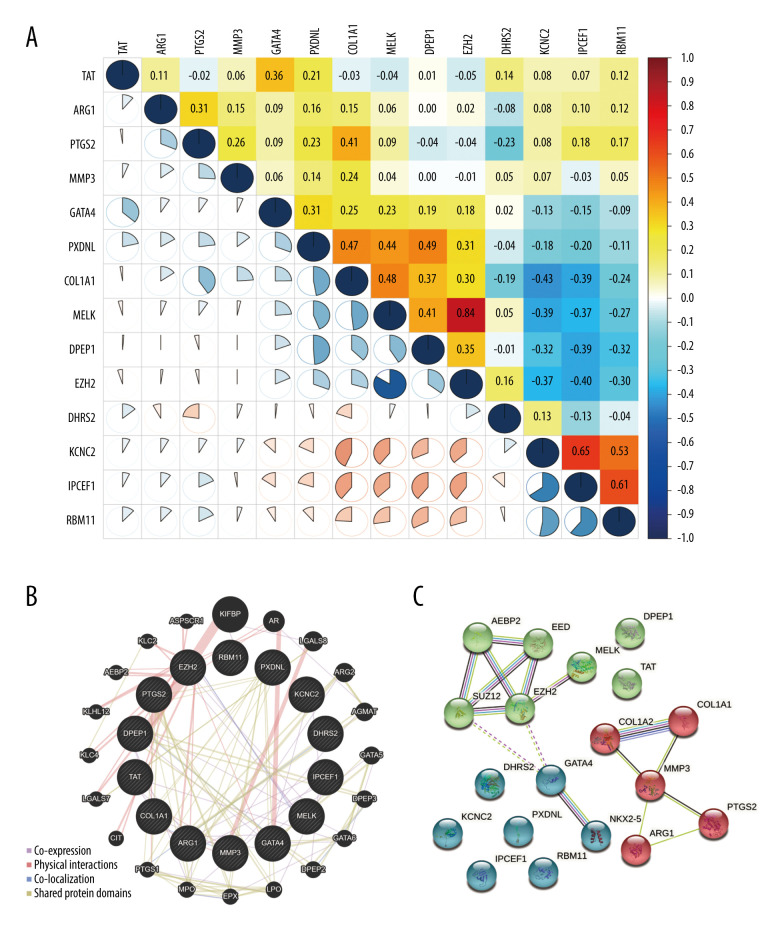
The correlations of the 14 prognosis-associated OSRGs were analyzed using Pearson’s correlation test (Figure 2A). The results showed that EZH2 was positively correlated with MELK (0.84). KCNC2 was positively correlated with IPCEF1 (0.65). RBM11 was positively correlated with IPCEF1 (0.61). The absolute values of other correlation coefficients were <0.5, which indicates that most of the prognosis-associated OSRGs were not highly correlated with each other.
Figure 2.
The relationship of prognosis-associated oxidative stress-related genes. (A) Pearson’s correlation analysis. Red indicates high correlation and blue indicates low correlation. (B) Gene interaction network. (C) Protein-protein interaction network. (R Studio, Version 1.2.5042, RStudio, Inc.).
A gene interaction network of OSRGs then was constructed in GeneMANIA and several types of network connections were identified, including co-expression, physical interactions, and shared protein domains (Figure 2B). The results demonstrated that the 14 OSRGs had complex and close interactions with KIFBP, AR, LGALS8, ARG2, AGMAT, GATA5, DPEP3, GATA6, DPEP2, LPO, EPX, MPO, PTGS1, CIT, LGALS7, KLC4, KLHL12, AEBP2, KLC2, and ASPSCR1.
In addition, a protein-PPI network was performed with STRING to explore the potential functional interaction among OSRGs (Figure 2C). They were clustered into 3 groups, based on K-means method. AEBP2, EED, SUZ12, EZH2 MELK, TAT, and DPEP1 were in cluster 1 (green). COL1A1, COL1A2, MMP3, PTGS2, and ARG1 were in cluster 2 (red). DHRS2, GATA4, NKX2-5, KCNC2, PXDNL, IPCEF1, and RBM11 were in cluster 3 (blue).
Immunohistochemical Analysis of Prognosis-Associated OSRGs
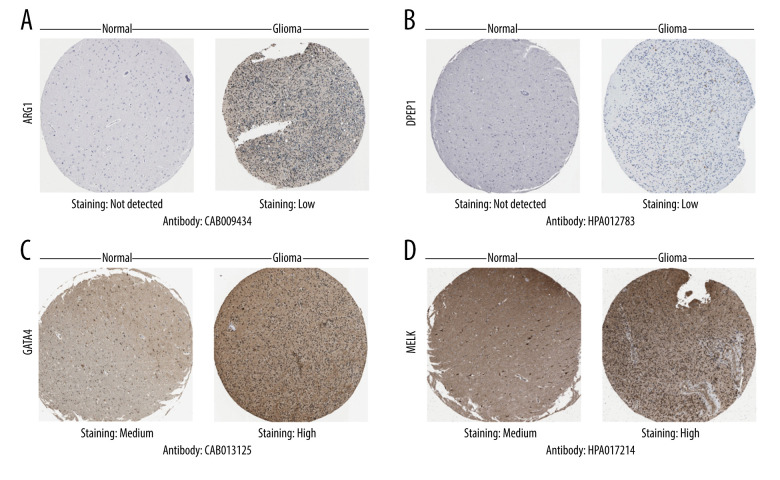
Next, we searched for each screened OSRGs in the HPA database. As was expected, the immunohistochemical images showed modifications in protein expression in glioma cells (Figure 3 and Supplementary Figure 1). Results of immunohistochemical staining obtained with the HPA database were basically consistent with the coefficient values in the risk score model. A positive coefficient value meant high expression of the corresponding protein in glioma that correlated with malignancy extent and poor prognosis, whereas a negative coefficient value meant the opposite. High or moderate staining was detected in EZH2, GTAT4, MELK, MMP3, and PTGS2. Low staining was observed in ARG1, COL1A1, DPEP1, and KCNC2. No staining was seen in DHRS2 or IPCEF1. In addition, the results also demonstrated that protein expression levels were increased in ARG1, DPEP1, EZH2, GTAT4, MELK, and PTGS2 compared with normal glial cells.
Figure 3.
Differential protein expression of oxidative stress-related genes in the Human Protein Atlas database. (A) ARG1 (CAB009434). (B) DPEP1 (HPA012783). (C) GATA4(CAB013125). (D) MELK (HPA017214). Normal: glial cells in normal tissue.
Risk Score Model Construction Based on Prognosis-Associated OSRGs
A risk score model was constructed, based on the obtained coefficient value and expression value of 14 prognosis-associated OSRGs. The equation was as follows:
| (2) |
Each gene’s coefficient value is accurate to 3 decimal places. The median risk score was 0.99. For clinical convenience, the value is approximated as 1.
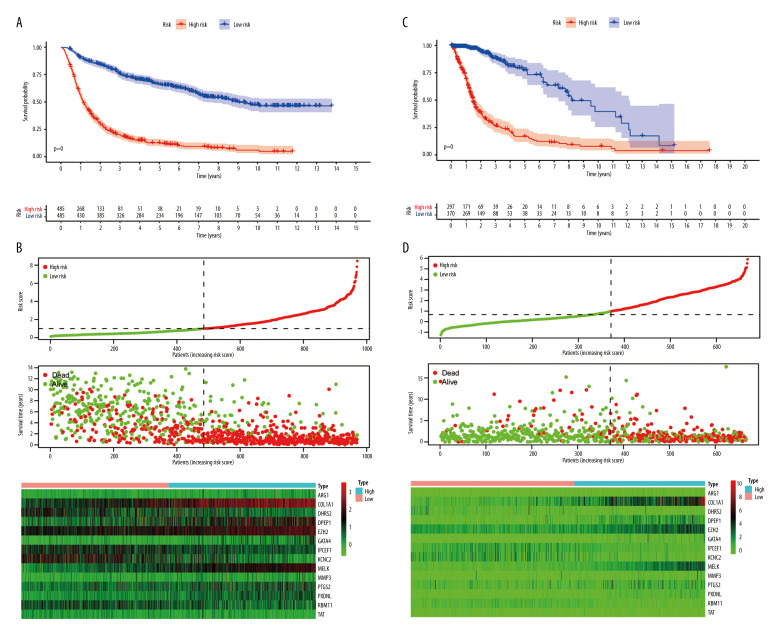
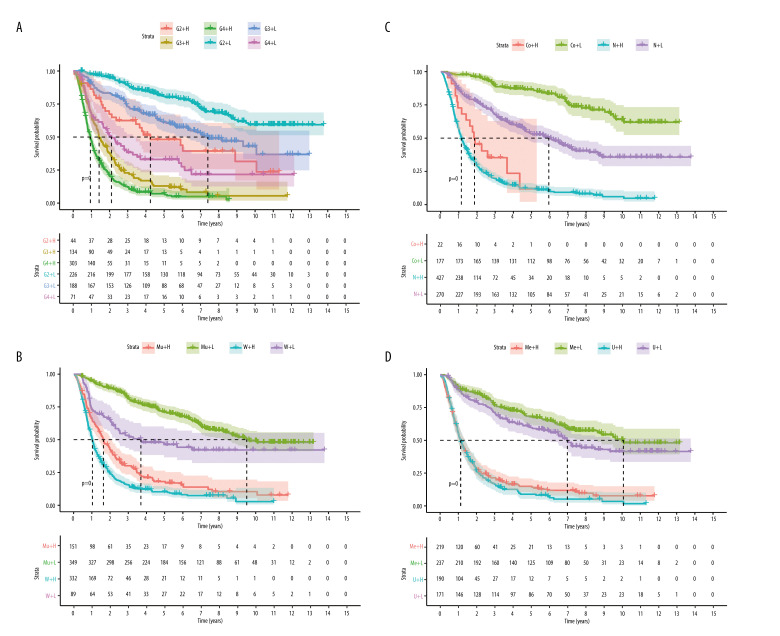
The glioma samples were divided into 2 groups according to their risk scores. Kaplan-Meier analysis (Figure 4A) demonstrated that the OS of the high-risk group was significantly shorter than for the low-risk group (P<0.001). The median OS of patients was 1.13 years in the high-risk group and 9.04 years in the low-risk group. The number of deaths progressively increased as the risk score increased (Figure 4B). This indicated that the risk score for OSRGs could accurately predict the survival of glioma patients. Kaplan-Meier analyses were also performed in different subtypes, including for grade and IDH mutation, 1p19q codeletion, MGMT promoter methylation, chemotherapy, and radiotherapy statuses (Figure 5 and Supplementary Figure 2). The results showed that high-risk patients in all subgroups had worse OS. High-risk patients with glioblastoma, IDH wild-type, without 1p19q codeletion or MGMT promoter methylation had the worst outcomes.
Figure 4.
Risk score model construction and external validation. (A) Kaplan-Meier survival curves for overall survival (OS) in the Chinese Glioma Genome Atlas (CGGA) cohort. (B) Risk score, survival status of patients, and heat map of the expression profile for the oxidative stress-related genes (OSRGs) in the CGGA cohort. (C) Kaplan-Meier survival curves for OS in The Cancer Genome Atlas (TCGA) cohort. (D) Risk score, survival status of patients and heat map of the expression profile for the OSRGs in TCGA cohort. (R Studio, Version 1.2.5042, RStudio, Inc.).
Figure 5.
Survival analysis of the Chinese Glioma Genome Atlas in different subgroups (H – high risk; L – low risk) (A) Grade (G – grade). (B) IDH mutation status (Mu – IDH mutation; W – wild-type). (C) 1p19q codeletion status (Co – 1p19q codeletion; N – no codeletion). (D) MGMTp methylation status (Me – MGMTp methylation; U – un-methylation). (R Studio, Version 1.2.5042, RStudio, Inc.).
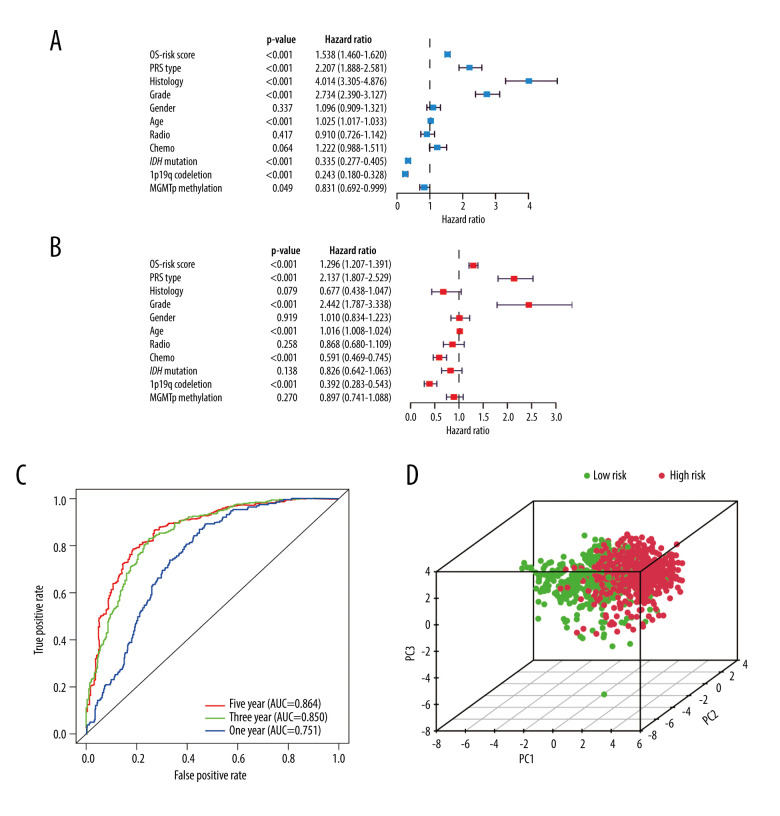
Independent Prognostic Value of the Risk Score Model
Excluding the samples with incomplete clinicopathological data, we performed further analyses on samples from 686 glioma patients (Table 2). The independent prognostic value of the risk score model was assessed with other clinicopathological parameters. Univariate (Figure 6A) and multivariate Cox regression analyses (Figure 6B) demonstrated that the risk score was correlated with the clinical prognosis of glioma patients (hazard ratio=1.296 in multivariate Cox regression, P<0.001). In the ROC analysis, the results revealed that the model had AUCs of 0.751 for 1-year survival, 0.850 for 3-year survival, and 0.864 for 5-year survival. All of the AUCs were >0.75 and had good predictive value. (Figure 6C).
Table 2.
Clinicopathological parameters of 686 glioma patients in the CGGA database.
| Parameters | Cases | Proportion (%) |
|---|---|---|
| Age (y) | ||
| 0–20 | 21 | 3.06 |
| 21–40 | 266 | 38.78 |
| 41–60 | 341 | 49.71 |
| 61–80 | 58 | 8.45 |
| Sex | ||
| Male | 399 | 58.16 |
| Female | 287 | 41.84 |
| Grade | ||
| WHO II | 177 | 25.80 |
| WHO III | 226 | 32.94 |
| WHO IV | 283 | 41.25 |
| Histology | ||
| A | 108 | 15.74 |
| AA | 160 | 23.32 |
| AO | 61 | 8.89 |
| O | 74 | 10.79 |
| GBM | 283 | 41.25 |
| PRS type | ||
| Primary | 442 | 64.43 |
| Recurrent | 218 | 31.78 |
| Secondary | 26 | 3.79 |
| Chemotherapy status | ||
| TMZ-treated | 501 | 73.03 |
| Untreated | 185 | 26.97 |
| Radiotherapy status | ||
| Treated | 544 | 79.30 |
| Untreated | 142 | 20.70 |
| IDH mutation status | ||
| Mutant | 371 | 54.08 |
| Wild-type | 315 | 45.92 |
| 1p19q codeletion status | ||
| Codeletion | 141 | 20.55 |
| Non-codeletion | 545 | 79.45 |
| MGMTp methylation status | ||
| Methylated | 386 | 56.27 |
| Unmethylated | 300 | 43.73 |
GBM – glioblastoma multiforme; TMZ – temozolomide; WHO – World Health Organization.
Figure 6.
Independent prognostic analysis in the Chinese Glioma Genome Atlas. (A) Univariate Cox regression analysis. (B) Multivariate Cox regression analysis. (C) Receiver operating characteristic curves for predicting 1-, 3-, and 5-year overall survival. (D) Principal component analysis. (R Studio, Version 1.2.5042, RStudio, Inc.).
In PCA analysis, all glioma samples were plotted onto a spatial coordinate system (PC1, PC2, PC3). Each dot represents a patient (Figure 6D). High-risk patients are represented by red dots and low-risk patients are represented by green dots. Relatively clear demarcations can be seen between the 2 groups of patients.
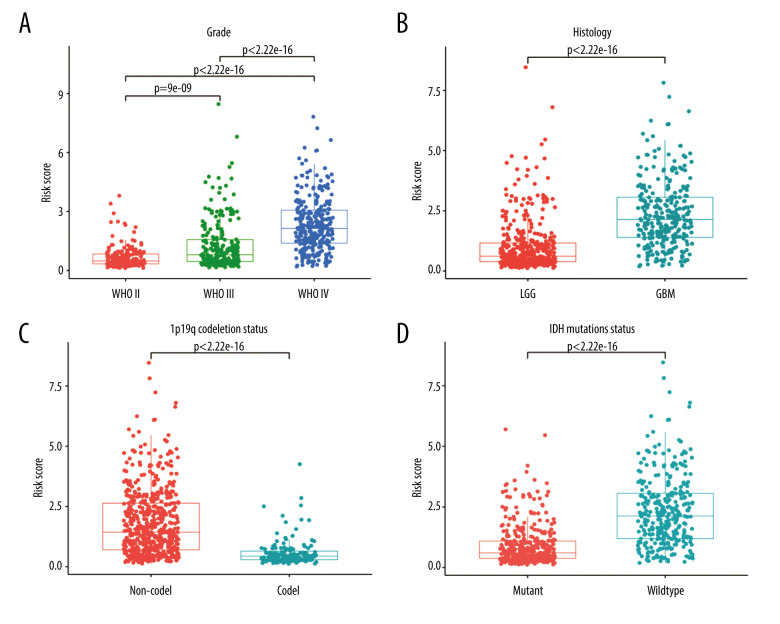
Relationship Between Risk Score and Clinic-Pathological Characteristics
To evaluate the relationship between risk score and other clinical characteristics, stratified analyses were performed. Significant differences in the risk score were observed based on stratification by age, grade, PRS type, histology type, and chemotherapy, 1p19q codeletion, IDH mutation, and MGMT promoter methylation statuses (Figure 7 and Supplementary Figure 1). When stratified by median age (43 years), the risk score was higher in older patients. When stratified by grade (Figure 7A), the risk score increased with the severity of the WHO classification. When stratified by PRS type, patients with recurrent glioma had higher risk scores. When stratified by histology (Figure 7B), GBM had significantly higher risk scores than LGG. When stratified by chemotherapy status, patients who received chemotherapy had higher risk scores, possibly because administration of chemotherapy was more frequent in high-risk glioma patients. Correspondingly, the risk score was higher in patients with no co-deficiency in 1p19q (Figure 7C), wild-type without IDH mutation status (Figure 7D), and methylation-free with MGMT promoter. No significant differences were detected in radiotherapy status. Similar results were also reported by Chen et al [30].
Figure 7.
Stratified analysis of risk scores based on different clinicopathological parameters. (A) Grade. (B) Histology. (C) 1p19q codeletion status. (D) IDH mutation status. (R Studio, Version 1.2.5042, RStudio, Inc.).
External Validation with TCGA Dataset
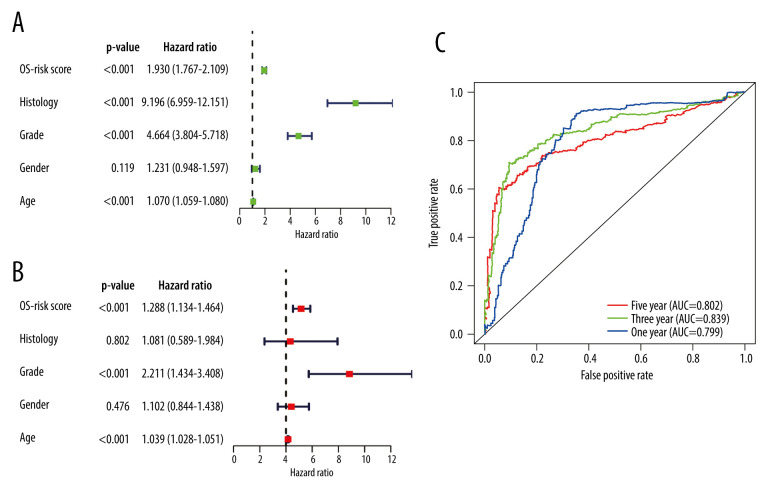
A total of 509 LGG patients and 159 GBM patients from the TCGA dataset were included in the present study. Based on the above risk score model, the risk score for each patient was calculated and the patients were divided into high- and low-risk groups based on the median (0.99) in the CGGA. Kaplan-Meier survival analysis showed a significant difference (P<0.001) between the groups (Figure 4C). The results showed that the number of deaths gradually increased as the risk score increased (Figure 4D). Univariate and multivariate Cox regression analyses (Figure 8A, 8B) demonstrated that the risk score was correlated with the clinical prognosis of glioma patients (hazard ratio=1.288 in multivariate Cox regression, P<0.001). ROC analyses revealed that the model had an AUC of 0.799 for 1-year survival, 0.839 for 3-year survival, and 0.802 for 5-year survival (Figure 8C). The results indicated that the risk score model could be generalized to other datasets.
Figure 8.
Independent prognostic analysis in The Cancer Genome Atlas: (A) Univariate Cox regression analysis. (B) Multivariate Cox regression analysis. (C) Receiver operating characteristic curves for predicting 1-, 3-, and 5-year overall survival. (R Studio, Version 1.2.5042, RStudio, Inc.).
Gene Set Enrichment Analysis
To analyze which pathways genes are mainly enriched in high- and low-risk groups in the CGGA cohort, GSEA was conducted to obtain cellular, metabolic, and functional annotation. We found that 137 of 170 gene sets were upregulated and 20 gene sets were significantly enriched in the high-risk group, at a nominal P<0.05. Thirty-three of 170 gene sets were upregulated in the low-risk group but none of them were significantly enriched (P>0.05).
In the high-risk group, the pathways of the top 20 gene sets were focal adhesion, p53 signaling, bladder cancer, cell cycle, small cell lung cancer, prostate cancer, ECM-receptor interactions, leukocyte trans-endothelial migration, natural killer cell-mediated cytotoxicity, complementary pathways, coagulation cascades, JAK-STAT signaling, cytokine-cytokine receptor interaction, pathways in cancer, DNA replication, mismatch repair, toll-like receptor signaling, pancreatic cancer, antigen processing and presentation, intestinal immune network for immunoglobulin A production, and homologous recombination.
In the low-risk group, the pathways for the top 20 gene sets were neuroactive ligand-receptor interaction, proximal tubule bicarbonate reclamation, taste transduction, long-term potentiation, cardiac muscle contraction, calcium signaling, alanine aspartate, glutamate metabolism, long-term depression, steroid hormone biosynthesis, amyotrophic lateral sclerosis, butanoate metabolism, biosynthesis of unsaturated fatty acids, olfactory transduction, phosphatidylinositol signaling system, type 2 diabetes mellitus, metabolism of xenobiotics by cytochrome p450, propanoate metabolism, gap junction, aldosterone-regulated sodium reabsorption, and pyruvate metabolism.
Through enrichment analysis, we found that the pathways enriched in the high-risk group are more related to multiple carcinomas and cancer-related pathways (Figure 9A, 9B). This obliquely points to the fact that glioma had a higher level of malignancy in the high-risk group. This further explains why OSRG expression level can be an effective prognostic predictor for glioma patients.
Figure 9.
Gene set enrichment analysis (GSEA) results in high-risk groups and a nomogram. (A) GSEA in multiple carcinomas (B) GSEA in multiple cancer-related pathways. (C) Nomogram for predicting 1-, 3- and 5-year survival rates in glioma patients. (R Studio, Version 1.2.5042, RStudio, Inc.).
Building Clinical Scoring Criteria with Nomogram
The nomogram scoring system was constructed using the risk score and other clinical parameters recorded in the CGGA clinical dataset (Figure 9C). With this scoring system, clinicians were able to calculate the survival rate at 1, 3, and 5 years for each patient and predict their prognosis effortlessly [31].
Notably, the length of each line in the nomogram scoring system represents the effects of different clinical parameters and their significance on the survival rate and prognosis. We observed that the risk score, histology classification, PRS type, and grade had a substantial impact on the survival rate. In comparison, sex and MGMT promoter methylation status had little impact on the prediction of prognosis. These results are consistent with a previous report [32].
Discussion
Glioma is a malignant brain tumor that arises within the brain parenchyma. Various molecular mechanisms of progression of these tumors and therapeutic strategies for them are being intensively studied. As early as 1997, the role of oxidative stress in glioma cells was reported in the literature [33]. The conclusion of that study was that H2O2 induces a marked intracellular acidosis that may influence the ability of glial cells. Undoubtedly, it began to unravel the mystery of oxidative stress in glioma. Numerous subsequent studies have shown that the role of oxidative stress in glioma is quite intricate. Glioma cell death, tumorigenesis [34], and drug resistance [35] are thought to be associated with oxidative stress.
Oxidative stress has been implicated in both cell survival and cell death. A transient increase in H2O2 promotes cell proliferation by reversibly inactivating several protein tyrosine phosphatases and the lipid phosphatase PTEN, which upregulate insulin/growth factor signaling and activity in mitogen-activated protein kinase (MAPK) [7]. Excess ROS induces apoptosis through loss of inner membrane permeability, disrupting membrane potential and resulting in the release of cytochrome C in mitochondria [36].
Intracellular ROS production is increased in glioma cells with a vigorous metabolic rate. To cope with this condition, multiple genes are involved in regulation of oxidative stress. For instance, PTPN2 was overexpressed in gliomas but can be strongly oxidated and inactivated by ROS (H2O2). PTPN2 deficiency promoted apoptosis of glioma cells in vitro [34]. GBM stores excess fatty acids with diacylglycerol-acyltransferase 1 (DGAT1). Dysfunction in DGAT1 resulted in excessive fatty acids moving into mitochondria for oxidation, which triggers accumulation of high levels of ROS, mitochondrial damage, and apoptosis [37]. NQO1 upregulation when PTEN was higher protected GBM from oxidative damage. Whereas overexpression of PINK1 repressed ROS by stabilizing antioxidative enzymes (NQO1, catalase, and SOD) and inhibited GBM cell proliferation [38]. Downregulating p53 by suppressing NF-κB signaling significantly reduced H2O2-induced glioma death [39]. Bcl-2 regulates H2O2-induces autophagy in glioma through Beclin 1 and Akt/mTOR signaling [16].
Oxidative stress is also closely related to the therapeutic strategy for glioma. Cadmium contributes to DNA fragmentation in glioma cells, which can be prevented with a cellular antioxidant, glutathione [40]. Increased activity of Apel/Ref-1 results in drug resistance to alkylating agents in glioma [35]. Camptothecin induces GBM apoptosis by synergizing with Fas activation via oxidative stress pathways [41]. Cannabidiol can contribute to glioma apoptosis through an oxidative stress mechanism by activating caspase-3 [42].
Because of the heterogeneity of glioma, there is a dire need for a method that can predict prognosis. To fully understand the role and importance of oxidative stress in glioma, we analyzed more than 400 OSRGs in the present study. We found 21 differentially expressed, disease-associated OSRGs from the CGGA database using the Wilcox signed-rank test. GO enrichment analysis revealed that they were mainly enriched in oxidative stress, redox reaction, as well as various lumens of the cell. KEGG enrichment analysis showed that they were mainly enriched in pathways associated with cancer, such as ECM-receptor interaction, IL-17 signaling, microRNAs in cancer, TNF signaling, PI3K-Akt signaling, biosynthesis of ubiquinone and other terpenoid-quinones, as well as the transcriptional misregulation in cancer. These results also underscore the complex role of OSRG genes in tumorigenesis and development.
Next, we assessed disease-associated genes into the context of glioma patients. Using univariate and multivariate Cox regression analyses, 14 prognosis-associated OSRGs were identified. To further explore the interrelationship of prognosis-associated genes, Pearson’s correlation test was performed. Most of the absolute values of the correlation coefficient were <0.8. With the help of GeneMANIA, we found that prognosis-associated OSRGs had complex and close interactions with KIFBP, AR, LGALS8, ARG2, AGMAT, GATA5, DPEP3, GATA6, DPEP2, LPO, EPX, MPO, PTGS1, CIT, LGALS7, KLC4, KLHL12, AEBP2, KLC2, and ASPSCR1. The PPI network analysis with STRING classified these genes into 3 distinct clusters. Immunohistochemical images from the HPA database showed that protein expression levels were increased in ARG1, DPEP1, EZH2, GTAT4, MELK, and PTGS2 compared with normal glial cells.
By reviewing the literature, we found that these prognosis-associated genes also play an important role in the development of a variety of carcinomas. A previous study demonstrated that overexpression of ARG1 enhances arginase activity, cell viability, migration, and invasion of Huh7 cells in vitro. It also can lead to the progression of hepatocellular carcinoma by upregulating the epithelial-mesenchymal transition process [43]. COL1A1 can bring about dermatofibrosarcoma protuberans and giant-cell fibroblastoma through fusion with the PDGFB gene [44]. DHRS2 is downregulated by HOXA13 and can contribute to gastric carcinogenesis via a p53-dependent pathway [45]. Dehydropeptidase-1 (DPEP1) promotes proliferation and survival of leukemia cells through activation of the pCREB pathway [46].
Ectopic expression of EZH2 in prostate cells can induce the transcriptional repression of a specific cohort of genes involved in prostate cancer [47]. GATA4 overexpression was shown to enhance the protective effect of cCFU-F-derived exosomes on myocardial ischemic injuries [48]. KCNC2 with tumor-specific alternative splicing is associated with the development of glioma [49]. The MELK pathway was shown to reduce drug resistance of maslinic acid in triple-negative breast carcinoma [50]. The stromal proteinase MMP3 is involved in mammary carcinogenesis with Str1 [51]. Stromal cells can express PTGS2 to maintain epithelial cell proliferation during colon injury [52]. PXDNL is also known as PMR1. Cells without a functional PMR1 gene accumulate intracellular manganese and are extremely sensitive to manganese ion toxicity [53]. The protein RBM11 is a tissue-specific splicing factor and has potential implications during neuron and germ cell differentiation [54]. The TAT protein of the HIV-1 virus can enter cells efficiently when added exogenously to tissue cultures and it can carry other molecules into cells [55].
We then constructed a risk score model with these OSRGs. We performed a Kaplan-Meier analysis and observed a significant difference in OS between the high- and low-risk groups. The patients in the high-risk group had a relatively shorter OS and half of them died within 1.13 years. Kaplan-Meier analyses also showed that high-risk patients in all subgroups had worse OS. ROC analyses also demonstrated that the risk score had independent predictive value in terms of 1-, 3-, and 5-year prognoses. External validation with a large sample of patients in TCGA cohort also confirmed the reliability of our model. Similar results were obtained through Kaplan-Meier survival, Cox regression, and ROC analyses.
After that, we performed a stratified analysis to assess the correlation between the risk score and other clinicopathological characteristics. The results revealed that the risk score differed significantly when stratified by age, grade, PRS type, histology type, and chemotherapy, 1p19q codeletion, IDH mutation, and MGMT promoter methylation statuses. We observed that the use of the different stratification schemes yielded different distributions of risk scores.
Through GSEA, we found that a large number of tumor-related pathways were enriched in the high-risk group. These correlations further revealed that the expression of OSRGs could be an effective predictor of prognosis in glioma patients and the complicated functions of OSRGs in these patients.
Finally, we developed a clinical scoring system for clinicians with a nomogram. It can be used to effectively estimate the survival rate for an individual patient at 1, 3, and 5 years. We found that some clinical parameters, such as sex and MGMT promoter methylation status, had little if any impact on the total scores. Some parameters, however, such as risk score, grade, histology, and PRS type, had a more substantial impact on the survival rate.
There are several limitations of the present study. First, the predictive power and molecular mechanism of OSRGs need further experimental validation. Second, the retrospective nature of the data may have resulted in some selection bias.
Conclusions
Oxidative stress strongly affects the generation, growth, proliferation, programmed death, and resistance to radiotherapy and chemotherapy of glioma. In the present study, we identified 14 key OSRGs associated with prognosis in glioma patients. The risk score model and nomogram created using these genes was shown to have good predictive ability. The role of these genes is worthy of further in-depth studies to provide new insights into the diagnosis and treatment of glioma.
Supplementary Data
Differential protein expression of oxidative stress-related genes in the Human Protein Atlas database. (A) PTGS2, (B) COL1A1, (C) DHRS2, (D) KCNC2, (E) EZH2, (F) MMP3, (G) IPCEF1.
Survival analysis of the Chinese Glioma Genome Atlas in different subgroups (H – high risk; L – low risk) (A) Chemotherapy status (Che – Chemotherapy; UC – No chemotherapy). (B) Radiotherapy status (Ra – Radiotherapy; UR – No radiotherapy). (R Studio, Version 1.2.5042, RStudio, Inc.).
Stratified analysis of risk scores based on different clinicopathological parameters. (A) Age. (B) PRS type. (C) Chemotherapy status. (D) MGMT promoter methylation status.
Footnotes
Conflict of interest: None declared
Declaration of Figures’ Authenticity
All figures submitted have been created by the authors who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: This work was supported by the National Natural Science Foundation of China (81702474, 81701329, and 81972351), the Department of Science & Technology of Shandong Province (ZR2019ZD33 and 2018CXGC1503), the Special Foundation for Taishan Scholars (ts20110814, tshw201502056, and tsqn20161067), the Jinan Science and Technology Bureau of Shandong Province (2019GXRC006), Clinical Research Center of Shandong University (No. 2020SDUCRCB002), Shandong Development and Reform Commission, and the Shandong Research Institute of Industrial Technology
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–46. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Patil N, Cioffi G, et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol. 2020;22(12 Suppl 2):iv1–96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131(6):803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairncross G, Jenkins R. Gliomas with 1p/19q codeletion: a.k.a. oligodendroglioma. Cancer J. 2008;14(6):352–57. doi: 10.1097/PPO.0b013e31818d8178. [DOI] [PubMed] [Google Scholar]
- 6.Everhard S, Kaloshi G, Criniere E, et al. MGMT methylation: A marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60(6):740–43. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38(2):167–97. doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golstein P, Kroemer G. Cell death by necrosis: Towards a molecular definition. Trends Biochem Sci. 2007;32(1):37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Degterev A, Huang Z, Boyce M, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1(2):112–19. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 10.Ortmann B, Druker J, Rocha S. Cell cycle progression in response to oxygen levels. Cell Mol Life Sci. 2014;71(18):3569–82. doi: 10.1007/s00018-014-1645-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balin AK, Fisher AJ, Carter DM. Oxygen modulates growth of human cells at physiologic partial pressures. J Exp Med. 1984;160(1):152–66. doi: 10.1084/jem.160.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterfield DA, Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci. 2019;20(3):148–60. doi: 10.1038/s41583-019-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaribeygi H, Sathyapalan T, Atkin SL, Sahebkar A. Molecular mechanisms linking oxidative stress and diabetes mellitus. Oxid Med Cell Longev. 2020;2020:8609213. doi: 10.1155/2020/8609213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Violi F, Loffredo L, Carnevale R, et al. Atherothrombosis and oxidative stress: Mechanisms and management in elderly. Antioxid Redox Signal. 2017;27(14):1083–124. doi: 10.1089/ars.2016.6963. [DOI] [PubMed] [Google Scholar]
- 15.Soon BH, Abdul Murad NA, Then SM, et al. Mitochondrial DNA mutations in grade II and III glioma cell lines are associated with significant mitochondrial dysfunction and higher oxidative stress. Front Physiol. 2017;8:231. doi: 10.3389/fphys.2017.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Kong X, Kang J, et al. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol Sci. 2009;110(2):376–88. doi: 10.1093/toxsci/kfp101. [DOI] [PubMed] [Google Scholar]
- 17.Kant S, Kesarwani P, Prabhu A, et al. Enhanced fatty acid oxidation provides glioblastoma cells metabolic plasticity to accommodate to its dynamic nutrient microenvironment. Cell Death Dis. 2020;11(4):253. doi: 10.1038/s41419-020-2449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pistollato F, Chen HL, Schwartz PH, et al. Oxygen tension controls the expansion of human CNS precursors and the generation of astrocytes and oligodendrocytes. Mol Cell Neurosci. 2007;35(3):424–35. doi: 10.1016/j.mcn.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Duan S, Li M, Wang Z, Wang L, Liu Y. H19 induced by oxidative stress confers temozolomide resistance in human glioma cells via activating NF-kappaB signaling. Onco Targets Ther. 2018;11:6395–404. doi: 10.2147/OTT.S173244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–83. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733–39. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu GC, Wang LG, Han YY, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Xu C, Yu Q, et al. Comprehensive landscape of STEAP family functions and prognostic prediction value in glioblastoma. J Cell Physiol. 2021;236(4):2988–3000. doi: 10.1002/jcp.30060. [DOI] [PubMed] [Google Scholar]
- 31.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–70. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
- 32.Han MZ, Huang B, Ni SL, et al. A validated prognostic nomogram for patients with newly diagnosed lower-grade gliomas in a large-scale Asian cohort. Neuro Oncol. 2020;22(5):729–31. doi: 10.1093/neuonc/noaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai KL, Wang SM, Chen CC, et al. Mechanism of oxidative stress-induced intracellular acidosis in rat cerebellar astrocytes and C6 glioma cells. J Physiol. 1997;502(Pt 1):161–74. doi: 10.1111/j.1469-7793.1997.161bl.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Wang F, Xu J, Chen Z. PTPN2 induced by inflammatory response and oxidative stress contributed to glioma progression. J Cell Biochem. 2019;120(11):19044–51. doi: 10.1002/jcb.29227. [DOI] [PubMed] [Google Scholar]
- 35.Silber JR, Bobola MS, Blank A, et al. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8(9):3008–18. [PubMed] [Google Scholar]
- 36.Orrenius S, Gogvadze V, Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem Biophys Res Commun. 2015;460(1):72–81. doi: 10.1016/j.bbrc.2015.01.137. [DOI] [PubMed] [Google Scholar]
- 37.Cheng X, Geng F, Pan M, et al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metab. 2020;32(2):229–42e8. doi: 10.1016/j.cmet.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo S, Lei K, Xiang D, Ye K. NQO1 is regulated by PTEN in glioblastoma, mediating cell proliferation and oxidative stress. Oxid Med Cell Longev. 2018;2018:9146528. doi: 10.1155/2018/9146528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta K, Babbar P, Srivastava T, et al. p53 dependent apoptosis in glioma cell lines in response to hydrogen peroxide induced oxidative stress. Int J Biochem Cell Biol. 2002;34(2):148–57. doi: 10.1016/s1357-2725(01)00106-6. [DOI] [PubMed] [Google Scholar]
- 40.Watjen W, Beyersmann D. Cadmium-induced apoptosis in C6 glioma cells: Influence of oxidative stress. Biometals. 2004;17(1):65–78. doi: 10.1023/a:1024405119018. [DOI] [PubMed] [Google Scholar]
- 41.Xia S, Rosen EM, Laterra J. Sensitization of glioma cells to Fas-dependent apoptosis by chemotherapy-induced oxidative stress. Cancer Res. 2005;65(12):5248–55. doi: 10.1158/0008-5472.CAN-04-4332. [DOI] [PubMed] [Google Scholar]
- 42.Massi P, Vaccani A, Bianchessi S, et al. The non-psychoactive cannabidiol triggers caspase activation and oxidative stress in human glioma cells. Cell Mol Life Sci. 2006;63(17):2057–66. doi: 10.1007/s00018-006-6156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You J, Chen W, Chen J, et al. The oncogenic role of ARG1 in progression and metastasis of hepatocellular carcinoma. Biomed Res Int. 2018;2018:2109865. doi: 10.1155/2018/2109865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon MP, Pedeutour F, Sirvent N, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15(1):95–98. doi: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 45.Han Y, Song C, Wang J, et al. HOXA13 contributes to gastric carcinogenesis through DHRS2 interacting with MDM2 and confers 5-FU resistance by a p53-dependent pathway. Mol Carcinog. 2018;57(6):722–34. doi: 10.1002/mc.22793. [DOI] [PubMed] [Google Scholar]
- 46.Zhang JM, Xu Y, Gale RP, et al. DPEP1 expression promotes proliferation and survival of leukaemia cells and correlates with relapse in adults with common B cell acute lymphoblastic leukaemia. Br J Haematol. 2020;190(1):67–78. doi: 10.1111/bjh.16505. [DOI] [PubMed] [Google Scholar]
- 47.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–29. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 48.Hao C, Lu Z, Zhao Y, et al. Overexpression of GATA4 enhances the antiapoptotic effect of exosomes secreted from cardiac colony-forming unit fibroblasts via miRNA221-mediated targeting of the PTEN/PI3K/AKT signaling pathway. Stem Cell Res Ther. 2020;11(1):251. doi: 10.1186/s13287-020-01759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung HC, Baggerly KA, Tsavachidis S, et al. Global analysis of aberrant pre-mRNA splicing in glioblastoma using exon expression arrays. BMC Genomics. 2008;9:216. doi: 10.1186/1471-2164-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Zhu X, Yin Y. Maslinic Acid enhances docetaxel response in human docetaxel-resistant triple negative breast carcinoma MDA-MB-231 Cells via regulating MELK-FoxM1-ABCB1 signaling cascade. Front Pharmacol. 2020;11:835. doi: 10.3389/fphar.2020.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternlicht MD, Lochter A, Sympson CJ, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98(2):137–46. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown SL, Riehl TE, Walker MR, et al. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117(1):258–69. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapinskas PJ, Cunningham KW, Liu XF, et al. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol Cell Biol. 1995;15(3):1382–88. doi: 10.1128/mcb.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedrotti S, Busa R, Compagnucci C, Sette C. The RNA recognition motif protein RBM11 is a novel tissue-specific splicing regulator. Nucleic Acids Res. 2012;40(3):1021–32. doi: 10.1093/nar/gkr819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fawell S, Seery J, Daikh Y, et al. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci USA. 1994;91(2):664–68. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential protein expression of oxidative stress-related genes in the Human Protein Atlas database. (A) PTGS2, (B) COL1A1, (C) DHRS2, (D) KCNC2, (E) EZH2, (F) MMP3, (G) IPCEF1.
Survival analysis of the Chinese Glioma Genome Atlas in different subgroups (H – high risk; L – low risk) (A) Chemotherapy status (Che – Chemotherapy; UC – No chemotherapy). (B) Radiotherapy status (Ra – Radiotherapy; UR – No radiotherapy). (R Studio, Version 1.2.5042, RStudio, Inc.).
Stratified analysis of risk scores based on different clinicopathological parameters. (A) Age. (B) PRS type. (C) Chemotherapy status. (D) MGMT promoter methylation status.