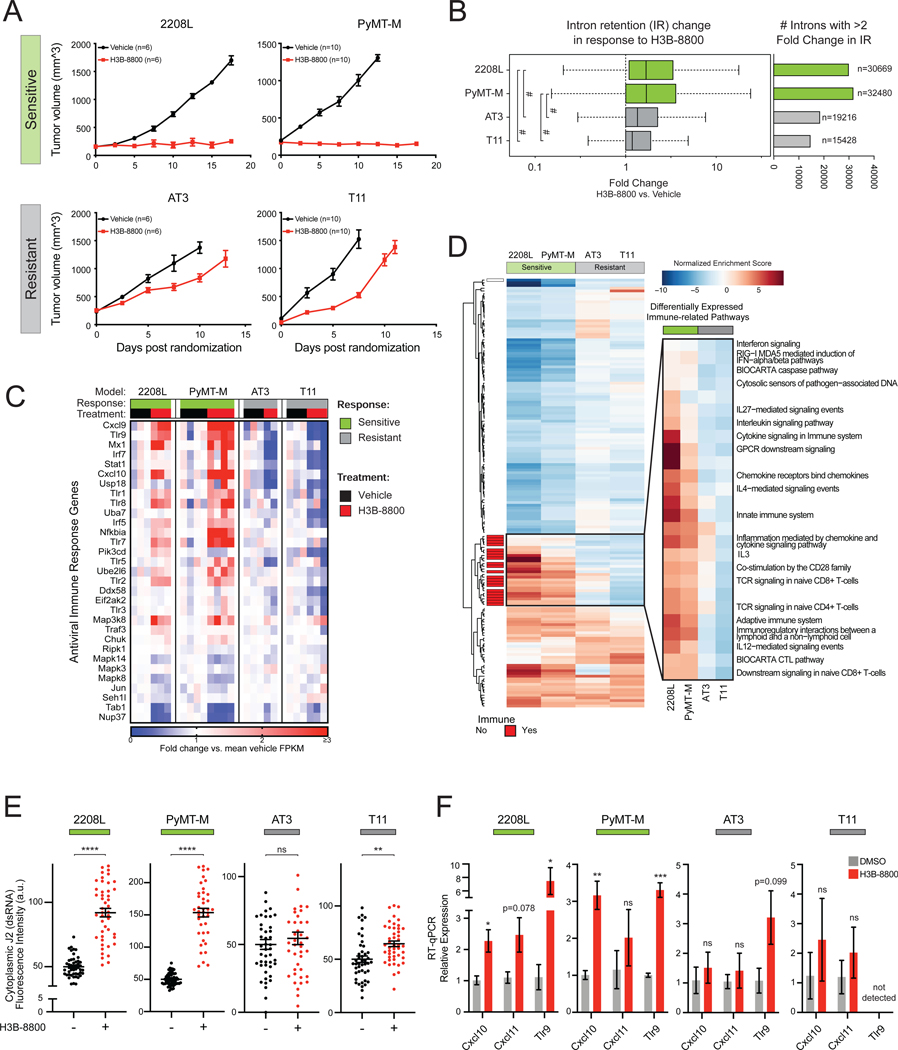
Figure 6. RNA Splicing Inhibition Induces Antiviral and Adaptive Signaling in Immune Competent Models of Breast Cancer.
(A) H3B-8800 impairs tumor progression heterogeneously across syngeneic murine TNBC tumor models. 2208L and PyMT-M tumor progression was significantly impaired (termed sensitive), while AT3 and T11 tumors progressed (termed resistant) (mean ± SEM number of animals plotted).
(B) H3B-8800 results in higher global intron retention in sensitive tumor models (p<2.2e-16, Mann-Whitney U). Boxplot (left) of transcriptome-wide IR scores. Bar plot (right) indicates number of introns with >2-fold change in IR in H3B-8800 vs. Vehicle-treated tumors.
(C) H3B-8800 stimulates expression of antiviral signaling genes in sensitive tumor models. Genes shown are part of KEGG and Reactome antiviral signaling-related pathways. Relative expression calculated as mean FPKM fold change vs. vehicle.
(D) Immune pathways are strongly induced by H3B-8800 in sensitive tumor models but not in resistant tumor models. Pathways shown from MSigDB C2 Canonical Pathways have GSEA FDR<0.05 in either both of the sensitive or both of the resistant models. Immune pathways (red) are annotated based on leading edge genes.
(E) Spliceosome inhibition leads to accumulation of cytoplasmic dsRNA in sensitive syngeneic models of TNBC in vitro. Cell lines derived from syngeneic mouse TNBC models were assessed for cytoplasmic dsRNA using J2-immunofluorescence. Quantification of cytoplasmic dsRNA signal intensity shown (mean ± SEM from ≥40 cells per group, two-tailed unpaired Student’s t-test).
(F) H3B-8800 induces transcriptional activation of antiviral immune signaling in sensitive syngeneic models of TNBC in vitro. Cell lines were treated with H3B-8800 and immune transcriptional activation was measured via RT-qPCR. Data are relative to DMSO (mean ± SEM, n=3 biological replicates, two-tailed unpaired Student’s t-test).
*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, #p<2.2e-16. See also Figure S6 and Table S3.