Figure 5. Accelerated glutamatergic gene and ELAVL4 expression precedes aberrant splicing in V337M neurons.
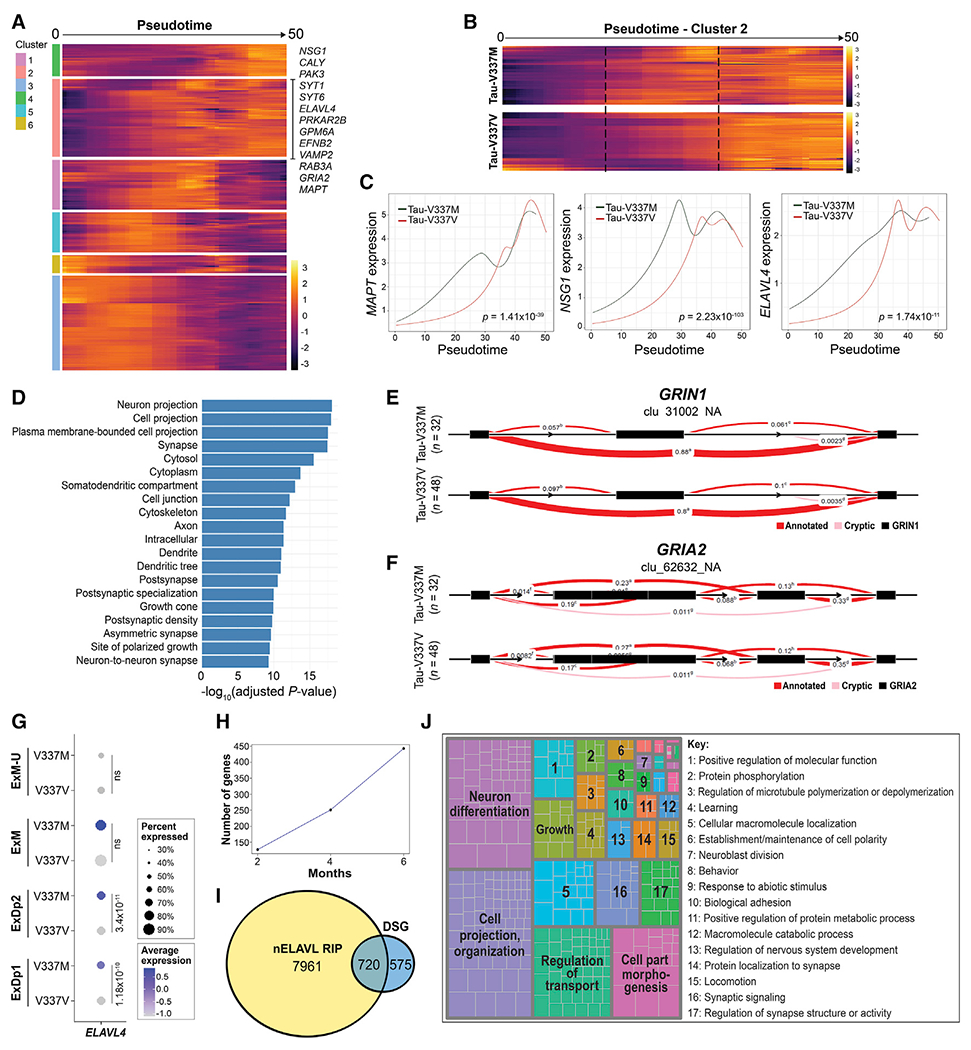
(A) Expression of genes with significantly differential trajectories over pseudotime in tau-V337M versus tau-V337V glutamatergic neurons by spline regression model. Trajectories grouped by unsupervised hierarchical clustering are centered across genes. Genes in significantly enriched glutamatergic signaling pathways are highlighted in cluster 2.
(B) Comparison of cluster 2 pseudotime trajectories in tau-V337M and tau-V337V glutamatergic neurons. Dashed lines highlight the central region of pseudotime, where gene expression differs between mutant and control cells.
(C) Trajectories of average MAPT, glutamatergic pathway gene NSG1, and ELAVL4 expression in tau-V337M and tau-V337V glutamatergic neurons over pseudotime. Adjusted p value for statistical comparison (spline regression) of trajectories is shown in the bottom right corner.
(D) GO pathways enriched for DSGs between 6-month-old tau-V337M and tau-V337V organoids.
(E and F) Leafcutter analysis of differentially spliced intron clusters in the glutamatergic receptor genes GRIN1 (E) and GRIA2 (F). Exons, black boxes. Red band thickness and inserted values represent proportion of spliced exon-exon pairs.
(G) Expression of ELAVL4 in tau-V337M and tau-V337V glutamatergic neurons. Dot size, proportion of cells expressing ELAVL4; depth of color, ELAVL4 expression level. Values: differential gene expression p value adjusted by MAST general linear model comparisons of differential expression.
(H) Number of known nELAVL gene targets by RIP (Scheckel et al., 2016) that are differentially spliced in tau-V337M organoids over time.
(I) Overlap between number of DSGs in tau-V337M organoids and genes in the brain known to be bound by nELAVL by RIP analysis (Scheckel et al., 2016).
(J) Semantic analysis of significant GO pathways enriched for DSGs known to be nELAVL targets (Scheckel et al., 2016).
See also Figure S6.
