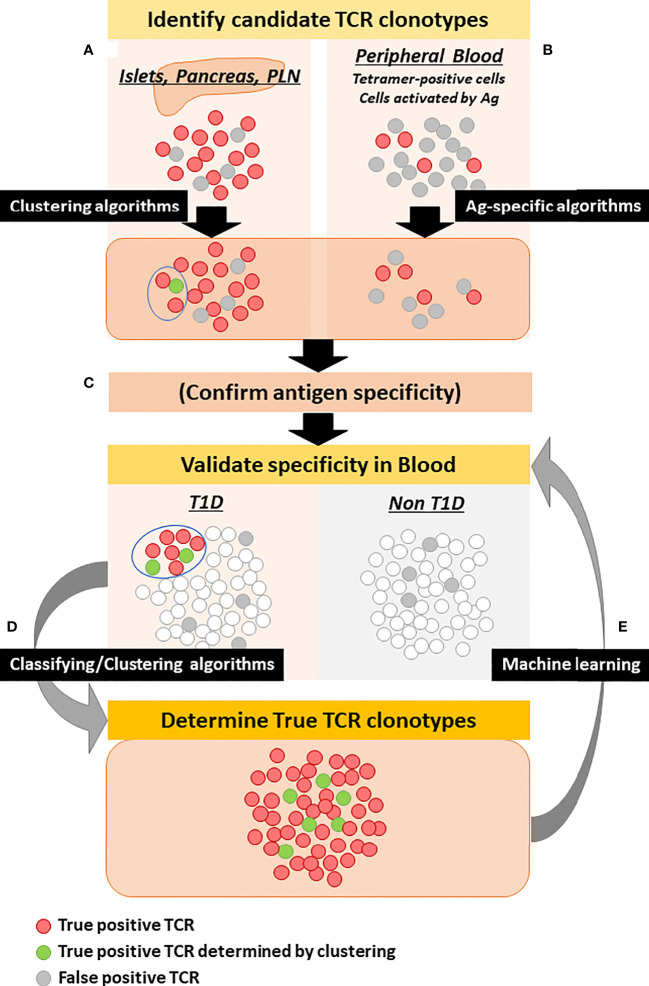
Figure 1.
Strategy to determine disease-specific TCR clonotypes. Red and gray circles represent true and false disease-specific TCR clonotypes, respectively. Green circles are true disease-specific clonotypes determined by clustering with known disease-specific TCR clonotypes. (A) TCRs detected in the islets, pancreata, and pancreatic lymph nodes, in particular those for which antigen specificity has been determined as well as those that are clustered with known disease-specific TCRs, can be the initial source for disease-specific TCR candidates. (B) TCRs detected from peripheral blood T cells enriched by antigen stimulation or peptide-MHC-conjugated multimers are also an initial source. Antigen-specific algorithms can enrich TCR clonotypes that are truly specific to antigens. (C) Candidate TCR clonotypes may be assessed for specificity to islet tissues, proteins, and peptides. (D) Using classifying algorithms, candidate TCR clonotypes are assessed for frequency in the blood of individuals with and without T1D to determine disease specificity. Simultaneously, clustering algorithms can select additional clonotypes that are clustered with known disease-specific TCR clonotypes. (E) TCR clonotypes selected by classifying and clustering algorithms are used for machine learning of antigen-specific algorithms to further determine true disease-specificity.