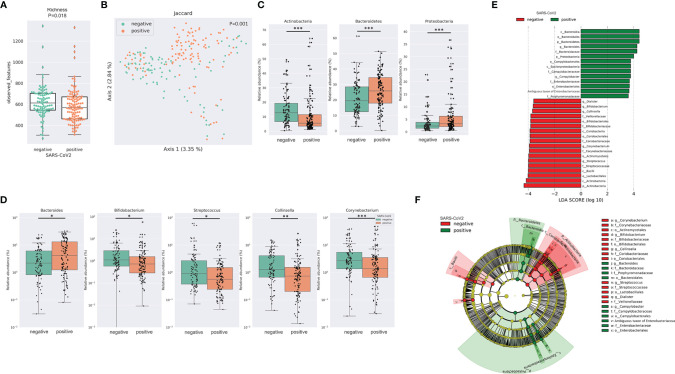
Figure 1.
SARS-CoV-2 infected patients exhibit distinct differences in the gut microbiome (A) Bacterial richness in SARS-CoV-2 positive and negative patients determined by observed features (amplicon sequence variants [ASV]). Kruskal-Wallis test was used to test for significant differences among groups. * FDR-P < .05; ** FDR-P < .01¸*** FDR-P < .001 (B) Principal Coordinates Analysis (PCoA) of SARS-CoV-2 positive performed with of Binary Jaccard distance matrix. PERMANOVA multivariate analysis was used to test for significant differences. (C) Differences in the relative abundance of bacterial phyla linked to SARS-CoV-2 infection. (D) Illustration of the differences in the relative abundance of bacterial genera linked to SARS-CoV-2 infection. (E) Linear discriminant effect size (LEfSe) analysis identified several bacterial taxa enriched in SARS-CoV-2 positive (green) and SARS-CoV-2 negative patients (red) (F) Cladogram reports the taxa showing different abundance values (LDA>3.5) according to LEfSe. Colors are presented in the color of the most abundant group (SARS-CoV-2 positive in green, SARS-CoV-2 negative in red).