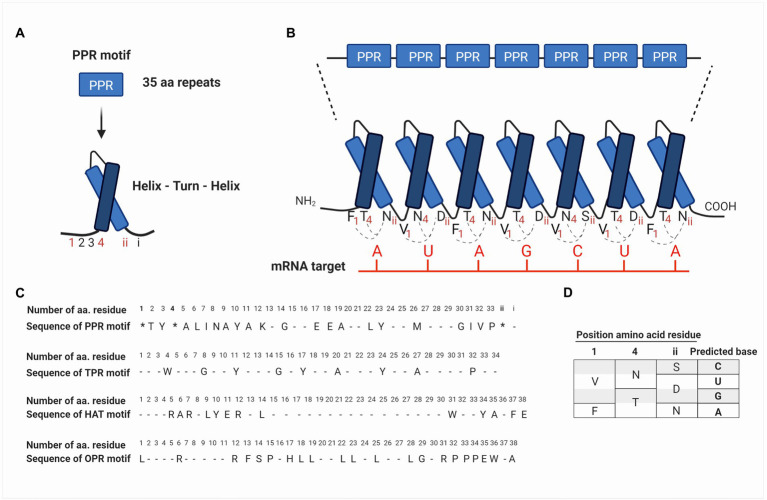
Figure 2.
Schematic representation of the structural arrangement of PPR proteins and their binding sites to mRNA targets based on the PPR code. (A) PPR proteins are composed of repeats of degenerate motifs of 35 amino acids. The PPR motif forms two antiparallel α-helices, which interact to produce a helix-turn-helix motif. Amino acids in the PPR motif are identified from 1 to 33 according to their position starting from the NH4-terminus. The last two amino acids are identified as i (last residue) and ii (penultimate residue), according to the position starting from the COOH-terminus (see also C in this figure). (B) PPR motifs are repeated in tandem and constitute a PPR protein, where each motif interacts with one ribonucleotide on the mRNA target. (C) The PPR motif identified for first time in Arabidopsis thaliana by Small and Peeters (2000) is shown at the top, followed by the consensus sequences of TPR and HAT motifs identified in proteins of Chlamydomonas reinhardtii, Physcomitrella patens, and A. thaliana (Bohne et al., 2016). At the bottom, the OPR motifs identified in TCB2 protein (Auchincloss et al., 2002), which contains the consensus sequence PPPEW present in most of OPR proteins is also shown. In the PPR motif, amino acid residues at positions 1, 4, and ii (indicated with an asterisk *) are determinants in the recognition of RNA bases. Conserved residues are indicated with the consensus one letter amino acid code, while degenerated residues are indicated with a dash. (D) Summary of the current PPR code proposed by Barkan et al. (2012) and Yagi and Shiina (2014). Different combinations of amino acids at positions 1, 4, and ii result in a code specific for a certain RNA base. For instance, Val (V), Asn (N), and Ser (S) at positions 1, 4, and ii, respectively, can bind cytosine in the mRNA target. Similarly, Phe (F), Thr (T), and Asn (N) bind adenine.