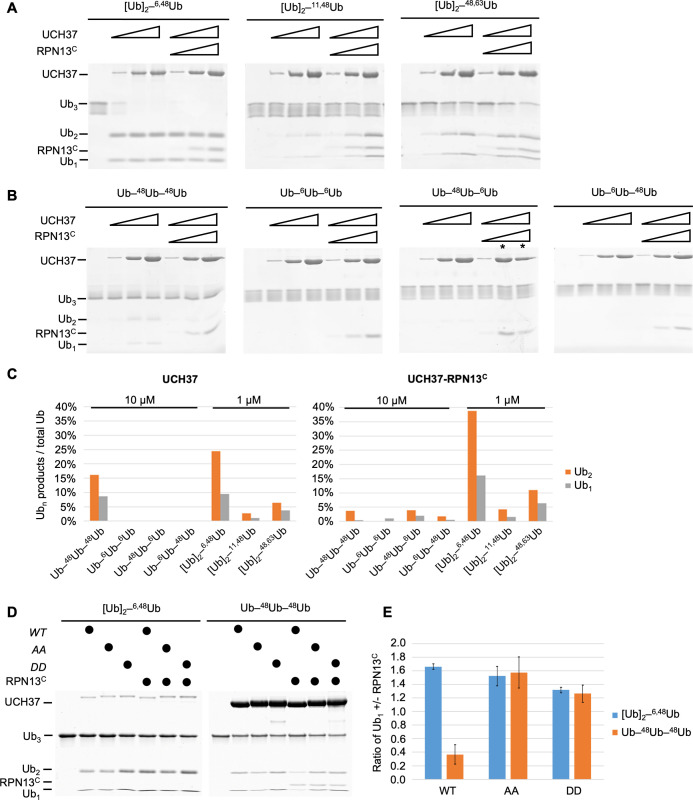
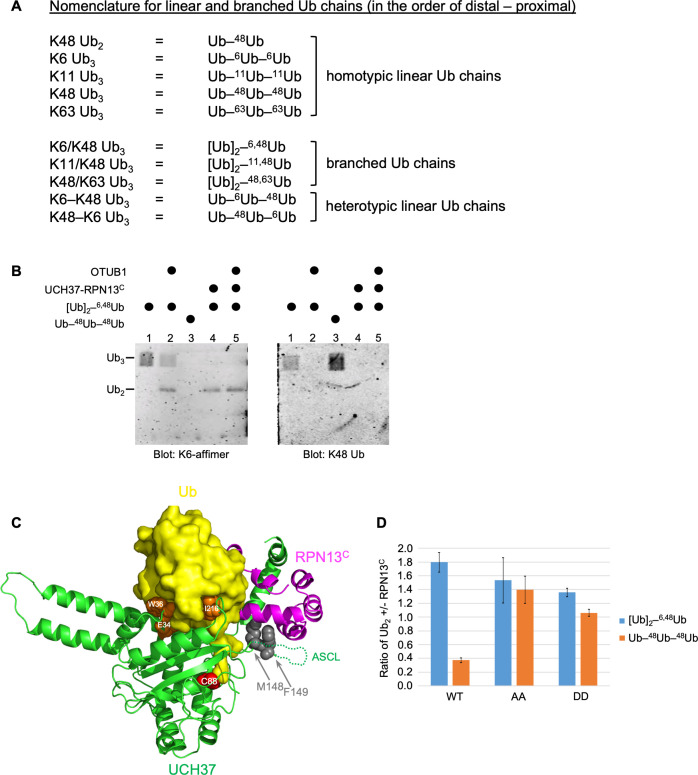
Figure 1. UCH37–RPN13C preferentially cleaves branched polyUb chains.
Branched (A) or linear (B) Ub3 substrates (5 μM) with the indicated Ub–Ub linkages were incubated for 1 hr at 37°C with 1, 5, or 10 μM His-TEV-UCH37, with or without the addition of equimolar RPN13C. Reactions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining. (C) Quantification of the Ub2 and Ub1 products from (A) and (B). For linear Ub3 substrates, results from incubations with 10 μM enzyme are shown; for branched Ub3 substrates, results from 1 μM enzyme are shown. (D) Wild-type (WT), M148A F149A (AA), or M148D F149D (DD) UCH37, alone or with the addition of equimolar of RPN13C, were incubated with Ub3 substrates for 2 hr at 37°C. Left, reactions contained 0.5 μM enzymes and 10 μM branched substrates. Right, reactions contained 10 μM enzymes and 5 μM linear Ub3. (E) Quantification from (D), plotted as the ratio of Ub1 produced in the presence over absence of RPN13C. Mean ± standard deviation (SD) from two independent replicates are shown.