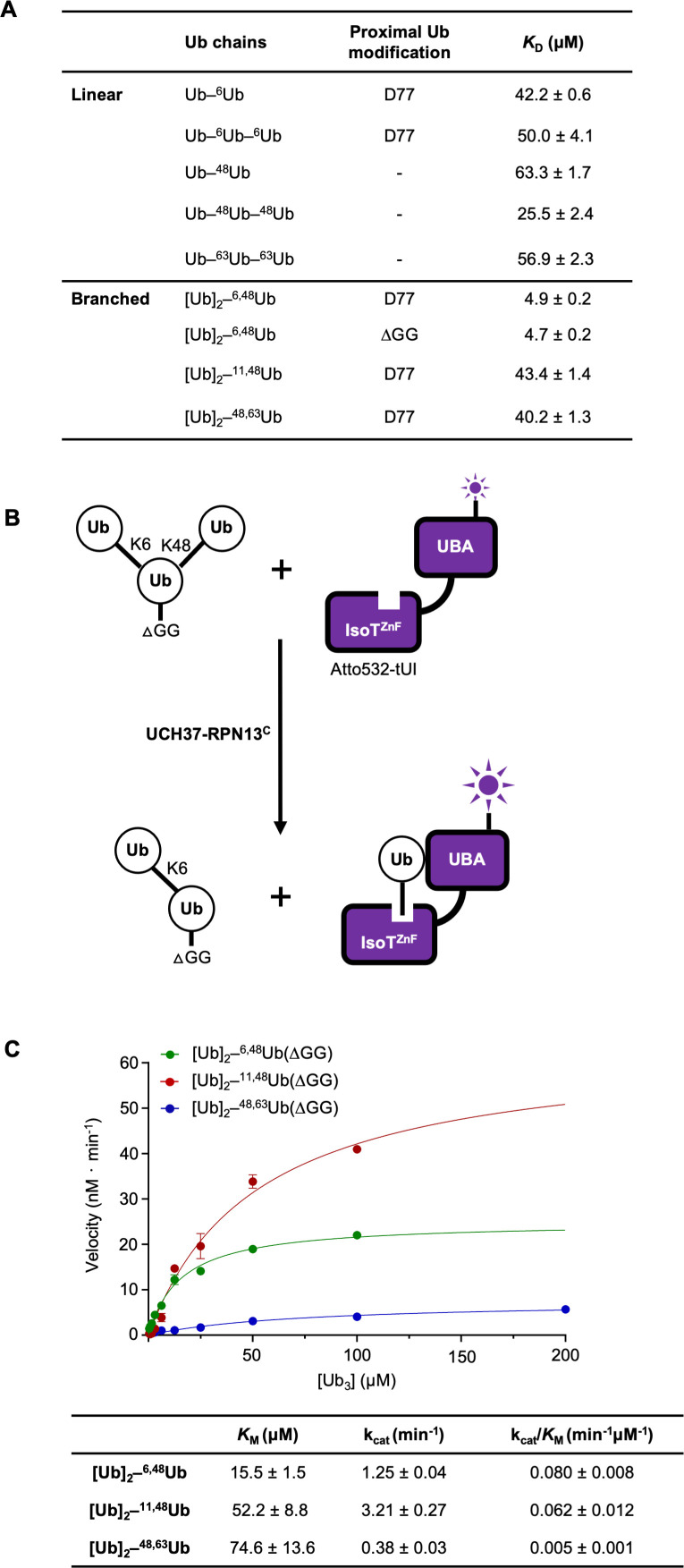
Figure 2. UCH37–RPN13C preferentially binds and deubiquitinates K6/K48-branched Ub3.
(A) Binding affinities between His-TEV-UCH37(C88S)–RPN13C and various polyUb chains were measured by microscale thermophoresis. Binding isotherms ( Figure 2—figure supplement 1) were fit with a single-site-binding model; best-fit KD values are shown with standard errors. (B) Schematic of the free Ub sensor-based deubiquitination assay. (C) Michaelis–Menten kinetics of branched Ub3 hydrolysis by NS–UCH37–RPN13C. The table shows best-fit KM, kcat, and kcat/KM values with standard errors from two independent replicates.