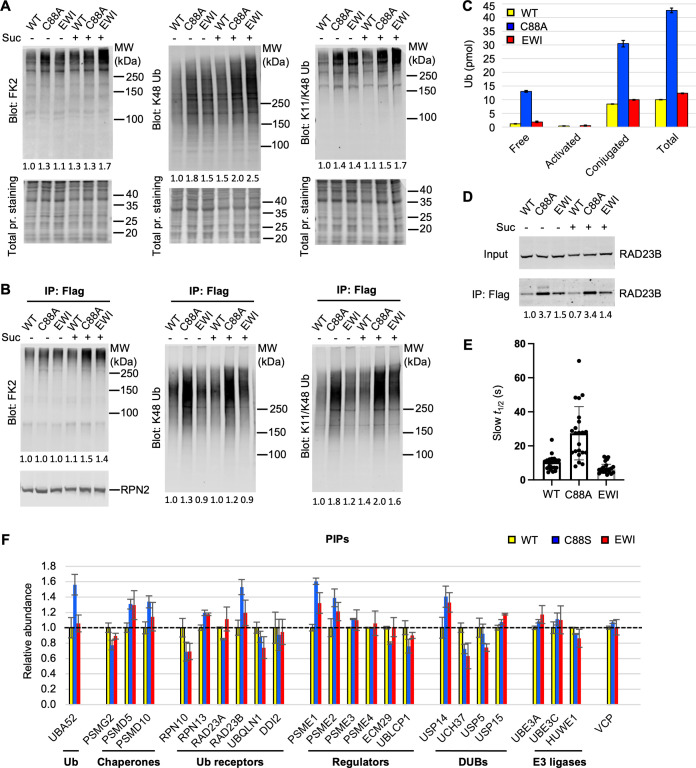
Figure 6. UCH37(C88A)-containing proteasomes accumulate polyUb species and RAD23B.
(A) Soluble cell lysates were collected from WT, C88A, and EWI cells with or without 30 min 0.2 M sucrose treatment, analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunblotting with indicated antibodies. Numbers indicate Ub signals relative to those in untreated WT cells after normalization against total protein stain signals. (B) UCH37-containing proteasomes were immunoprecipitated and analyzed as described in (A). Numbers below the lanes indicate Ub signals relative to those in untreated WT cells after normalization against RPN2 signals. (C) Different types of Ub species associated with UCH37-containing proteasomes immunoprecipitated from HEK293 cells were quantified by a free Ub sensor-based assay (Choi et al., 2019). Shown are mean ± SD from two independent replicates. (D) RAD23B accumulation detected on UCH37(C88A)-containing proteasomes as described in (A) and (B). (E) Fluorescence recovery after photobleaching (FRAP) analysis of GFP-UCH37 after 0.2 M sucrose treatment demonstrates that C88A-containing proteasomes are less mobile. At least 20 foci from 7 or 8 cells were analyzed from each cell line. After fitting the FRAP curve with two exponentials, the slow t1/2 of each focus were plotted with mean and SD indicated. (F) UCH37-containing proteasomes from WT, C88S, and EWI cells were isolated with immobilized anti-GFP nanobodies, trypsin digested, and the peptides analyzed by tandem mass tag (TMT) mass spectrometry. Protein abundances were measured from two independent pulldowns, each analyzed by mass spectrometry in triplicate, and plotted as mean ± coefficient of variation (CV) after normalizing against signals from WT samples.