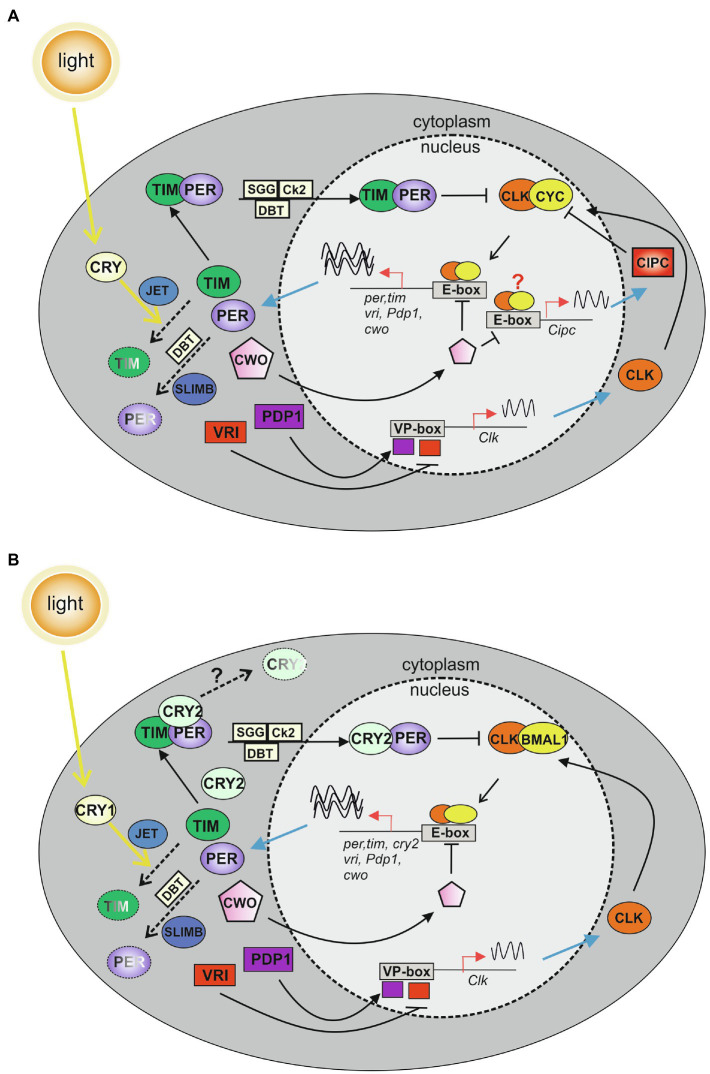
Figure 2.
Model of the major transcription–translation feedback loops (TTLs) in the circadian molecular clock of Drosophila melanogaster (A) and Lepidoptera (B). (A) In the first TTL of D. melanogaster, CLOCK (CLK) and CYCLE (CYC) dimerize and bind to E-boxes in the promoters of per and tim clock genes. PERIOD (PER) and TIMELESS (TIM) enter the nucleus as a complex, and inhibit CLK-CYC activity (Allada et al., 1998; Bae et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Glossop et al., 1999; Lee et al., 1999). A second TTL modulates Clk expression: CLK:CYC dimers induce the transcription of vrille (vri) and par domain protein 1 (Pdp1) genes. VRI and PDP1 compete for the same element (VP-box) in the Clk promoter, controlling Clk transcription (Cyran et al., 2003). In the third TTL, CLK:CYC dimers activate the transcription of clockwork orange (cwo). CWO enters the nucleus and inhibits CLK:CYC activity by binding to E-box elements of clock genes (Kadener et al., 2007; Lim et al., 2007; Richier et al., 2008). CWO acts also indirectly promoting CLK:CYC-mediated transcription, by repressing the production of the CLK:CYC inhibitor CLOCK INTERACTING PROTEIN CIRCADIAN (CIPC; Rivas et al., 2021). Cipc transcription modulation might occur at the level of the E-boxes detected in the Cipc locus (red question mark; Rivas et al., 2021). Phosphorylation mediated by DOUBLETIME (DBT), CASEIN KINASE 2 (CK2), and SHAGGY (SGG) modulate clock protein activities, regulating protein–protein interactions, nuclear translocation, and degradation (Curtin et al., 1995; Saez and Young, 1996; Price et al., 1998; Rothenfluh et al., 2000; Martinek et al., 2001). Light activates the photoreceptor CRYPTOCHROME (CRY), which interacts with TIM and mediates its degradation in combination with JETLAG (JET). SUPERNUMERARY LIMBS (SLIMB) signals PER degradation (Emery et al., 1998; Stanewsky et al., 1998; Lin et al., 2001; Koh et al., 2006). (B) The first lepidopteran TTL includes CLK and BMAL1, which dimerize and bind to the E-boxes in the promoters of per, tim, and cry2 genes. In the cytoplasm, PER, TIM, and CRY2 form a complex. CRY2, stabilized by PER, enters into the nucleus to inhibit CLK:BMAL1 activity (Zhu et al., 2005, 2008; Yuan et al., 2007). The second and third TTLs are speculative and include lepidopteran orthologs of Drosophila core and regulatory clock components identified in lepidopteran assembled genomes (Zhan et al., 2011; Derks et al., 2015). In the second TTL, the CLK:BMAL1 complex induces the transcription of vri and Pdp1 genes, which, once translated, compete for the same element (VP-box) in the Clk promoter, controlling Clk transcription. In the third TTL, a CLK:BMAL1 dimer activates the transcription of cwo. CWO enters the nucleus and inhibits CLK:BMAL1 activity by binding to E-box elements of clock genes. Phosphorylation mediated by DBT, CK2, and SGG modulate clock protein activities, regulating protein–protein interactions, nuclear translocation, and degradation. Light is responsible for TIM degradation in a process mediated by CRY1 and JET. SLIMB is involved in PER degradation. Pathways triggering to CRY2 degradation are still unknown (black question mark). Dashed arrows indicate degradation pathways; sinusoidal lines show transcription activity.