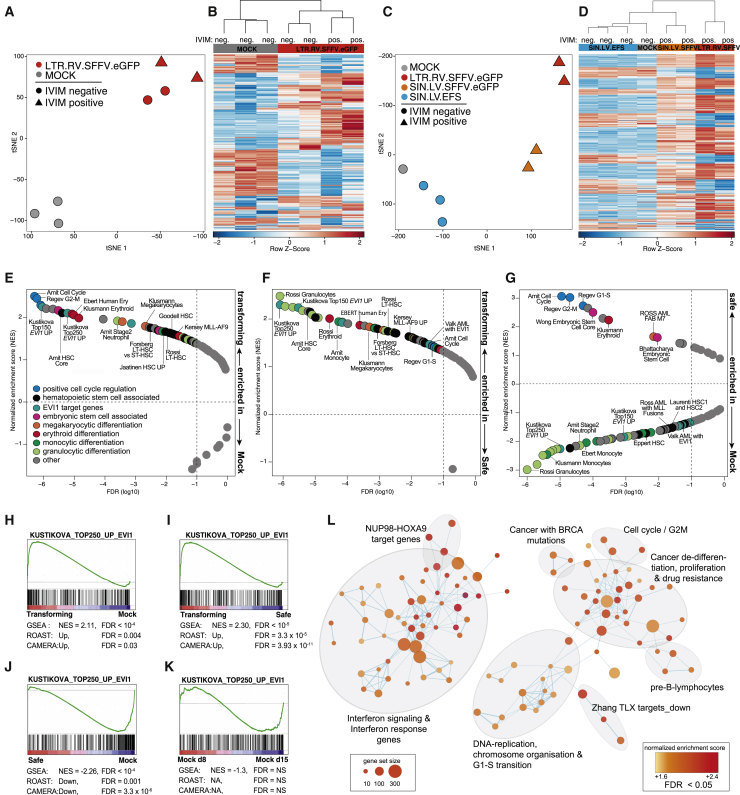
Figure 2.
Transforming vectors impose an oncogenic gene expression signature in murine HSPCs
(A) t-distributed stochastic neighbor embedding (t-SNE) of three mock samples (gray) and 4 samples transduced with LTR.RV.SFFV (red) from one SAGA assay (ID 120411) using all 36,226 annotated probes. (B) Hierarchical clustering of the samples shown in (A) based on the most variable probes (top 1%). (C) t-SNE of a second SAGA assay (ID 150128, 36,226 annotated probes). (D) Hierarchical clustering of the samples shown in (C) based on the most variable probes (top 1%). (E) Gene set enrichment analysis (GSEA) of hematopoiesis-associated gene sets (Table S3, tab 1) of samples transduced with IVIM-transforming vectors versus mock controls. Plotted are normalized enrichment scores (NESs) against the false discovery rate (FDR). The enrichment cutoff (FDR < 0.1) is indicated by the dashed line. (F) GSEA of IVIM-transforming vectors against IVIM-safe vectors. (G) GSEA of samples transduced with IVIM-safe vectors against mock controls. (H–K) GSEA plots for EVI1 target genes20 for the contrasts (H) transforming versus mock, (I) transforming versus safe, (J) safe versus mock, (K) mock day 8 versus mock day 15. (L) Enrichment map of highly upregulated (FDR < 0.005) gene sets from MSigDB in samples transduced with transforming vectors compared with mock control and safe samples.