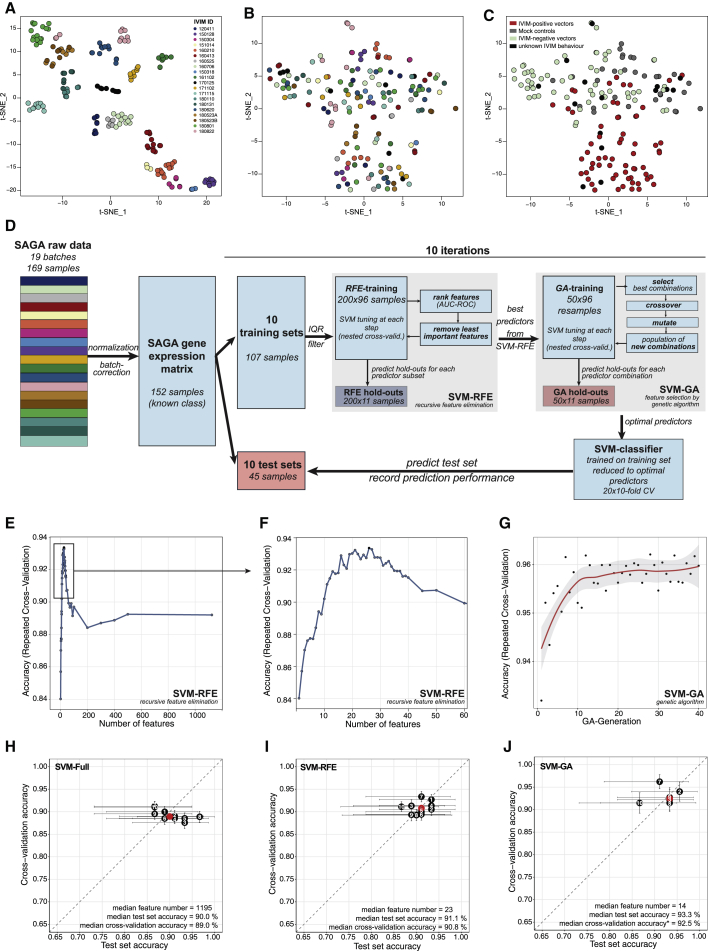
Figure 3.
Development phase of an SVM classifier to predict genotoxicity
(A–C) Data preprocessing. (A) t-SNE representation of all 169 SAGA assays after quantile normalization using all 39,428 probes. The coloring scheme encodes individual SAGA assays. (B) t-SNE of the 169 SAGA-samples after quantile normalization and ComBat correction using the same color key as in (A). (C) t-SNE plot as in (B) with the samples color coded according to vector properties in the IVIM assay. IVIM positive, transforming vectors; IVIM negative, nontransforming vectors; mock, untransduced controls; unknown, IVIM data inconclusive. (D) Scheme of classifier development during the development phase. The complete raw dataset was quantile normalized and batch corrected. The dataset was split 10 times into training (70% of samples) and test sets (30% of samples). Feature selection by SVM-RFE and SVM-GA was performed by further splitting the training sets using repeated cross-validation and monitoring prediction performance using the hold-out samples. Tuning of the SVM was performed at each step of the feature selection routines using nested cross-validation. An SVM with radial kernel was trained on the training set reduced to the optimal predictors found by SVM-RFE and SVM-GA and used to predict the test set. (E and F) Performance profile of SVM-RFE: accuracy on the hold-out samples plotted against the number of remaining probes during SMV-RFE for a representative training set (split 7). (G) Performance profile of SVM-GA: accuracy on the hold-out samples plotted against generation of the GA for training set 7. (H and I) Estimates of the prediction accuracy for the full models (H), RFE models (I), and GA models (J) using the test set (x axis) or repeated cross-validation (y axis). The horizontal and vertical bars represent the 95% confidence intervals using the test set and resampling approach, respectively.