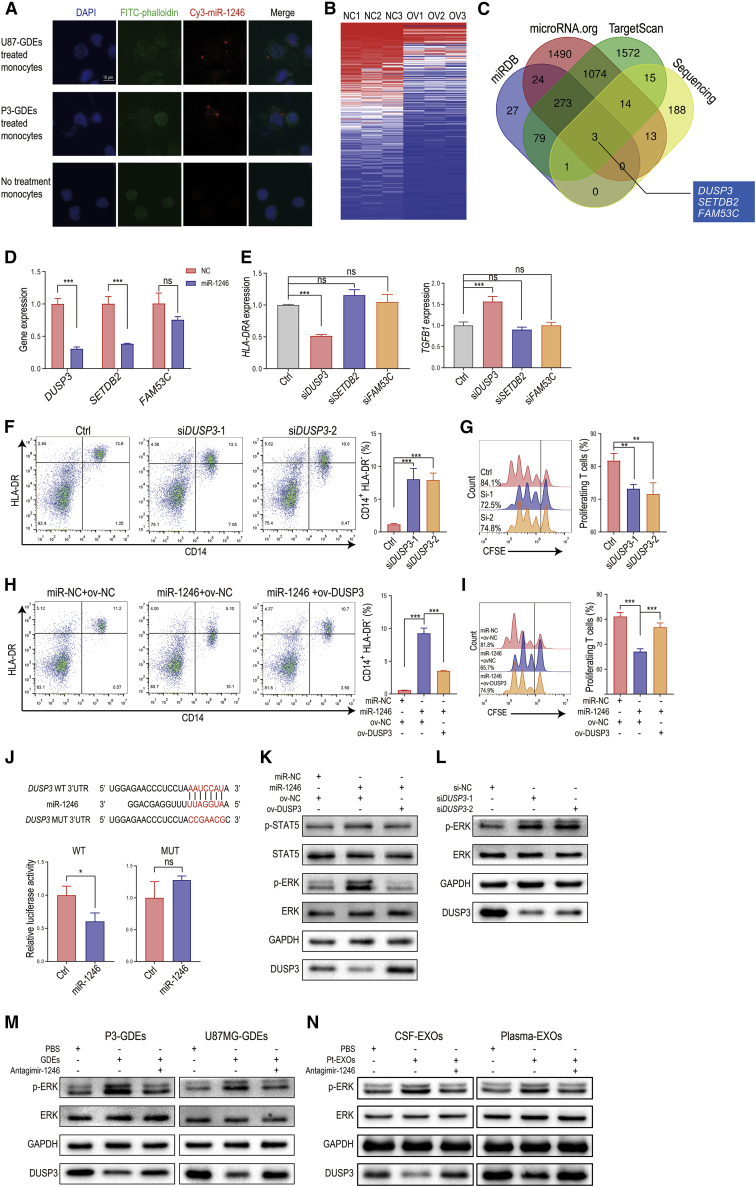
Figure 3.
miR-1246 in GDEs promotes MDSC differentiation and function through the DUSP3/ERK pathway
(A) GDEs derived from Cy3-tagged miR-1246-expressing glioma cells were used to treat human monocytes for 48 h. The monocytes were observed with confocal microscopy. Scale bar, 10 μm. (B) Heatmap of RNA sequencing results for miR-1246-overexpressing and control U937 cell lines. (C) Venn diagram identifying the overlap of miR-1246 target genes identified with the miRDB, microRNA.org, and TargetScan databases and the miR-1246 overexpression sequencing results. (D) DUSP3, SETDB2, and FAM53C expression was studied in miR-1246-overexpressing human CD14+ monocytes. (E) HLA-DRA and TGFB1 expression levels were measured after DUSP3, SETDB2, or FAM53C knockdown in monocytes. (F) The percentage of CD14+HLA-DR- MDSCs among DUSP3 knockdown PBMCs was measured by flow cytometry. (G) MDSCs with DUSP3 knockdown and control MDSCs were co-cultured with human CD8+ T cells. The proliferation of CD8+ T cells was measured. (H and I) The differentiation and function of MDSCs induced by miR-NC or miR-1246 mimics and nonsense sequence or DUSP3 overexpression plasmids. (J) Schematic diagram of the predicted wild-type binding site between miR-1246 and the DUSP3 3′ UTR and the mutated sequences. (K and L) Activation of the ERK and STAT5 pathways was studied after miR-1246 overexpression and DUSP3 knockdown. (M) GDE stimulation increased the phosphorylation of ERK, and this process was blocked by antagomir-1246. (N) Glioma patient CSF- and plasma-isolated exosomes activate the DUSP3/ERK pathway, and this effect was blocked by antagomir-1246. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.