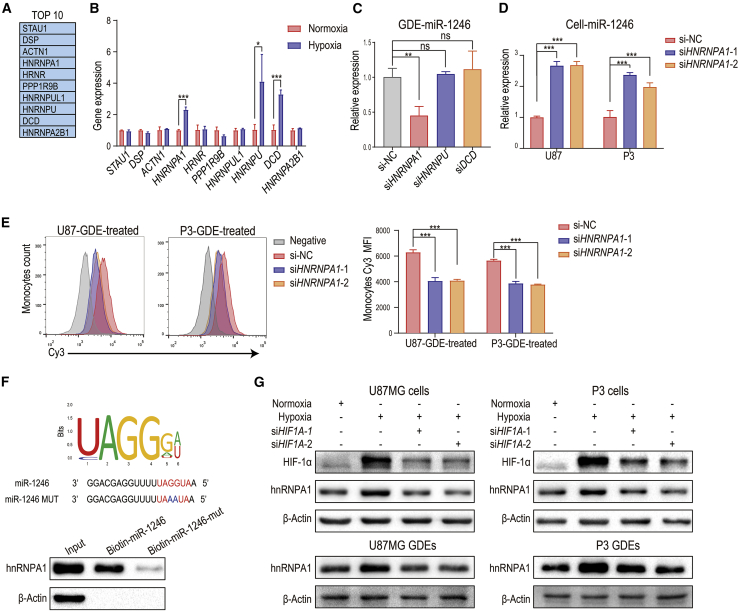
Figure 6.
Hypoxia-inducible hnRNPA1 selectively packages miR-1246 into exosomes
(A) In the pull-down assay,21 biotinylated miR-1246 was pulled down with protein lysates from wild-type (WT) HCT116 cells. Proteins pulled down with miR-1246 were analyzed with mass spectrometry. The top 10 identified proteins are displayed in the table. (B) The expression of the top 10 proteins in the pull-down assay was examined by quantitative real-time PCR in hypoxia- and normoxia-stimulated U87MG glioma cells. (C) The expression of miR-1246 in hypoxia-stimulated HNRNPA1, HNRNPU, and DCD knockdown and control U87MG GDEs was examined. (D) The expression of miR-1246 was examined in control and HNRNPA1 knockdown hypoxia-stimulated glioma cells. (E) Monocytes were treated with GDEs derived from Cy3-miR-1246-overexpressing U87 and P3 cells transfected with NC or si-HNRNPA1. The mean Cy3 fluorescence intensity of treated monocytes was measured. (F) The interaction between hnRNPA1 and miR-1246 was validated by pull-down assays. The binding capacity of hnRNPA1 to miR-1246 was impaired when UAGGUA was mutated to UAAAUA. (G) The expression of hnRNPA1 was studied in normoxia- and hypoxia-stimulated U87MG and P3 cells and GDEs. HIF1A knockdown impaired hypoxia-induced hnRNPA1 expression. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.