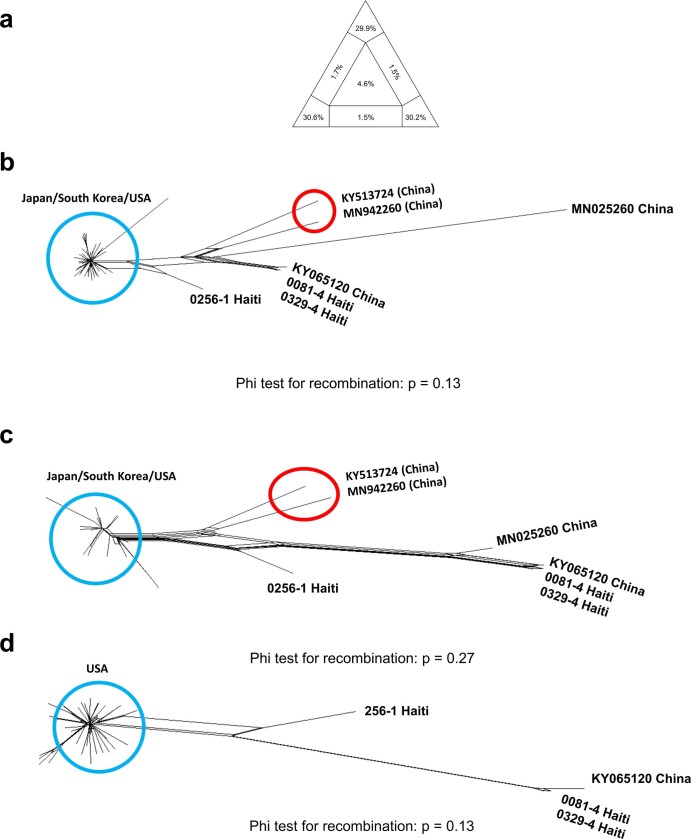
Extended Data Fig. 2. Assessment of phylogenetic signal and Neighbor net plot of PDCoV fragments.
a, Likelihood mapping of the final 109 sequences alignment (see Methods) showing extremely low phylogenetic noise (4.6% in the center of the triangle), as required for reliable phylogeny inference. b, Neighbor net inferred from pair-wise p-distances of 47 genomes for the major genome fragment identified by RDP4 (see Methods). Recombination signal was assessed by the PHI test. No significant evidence of recombination was found in either fragment (PHI test p > 0.05). c, Neighbor net and PHI test for the minor genome fragment identified by RDP4 (see Methods). The only change in topology, in the split decomposition networks shown in panels a and b, concerned two Chinese sequences (highlighted by the red circle) displaying mixed ancestry possibly due to homoplasy. These sequences were also removed before performing any further phylogenetic analyses. d, Neighbor net and PHI test for the full genome alignment only including Haitian Hu-PDCoV strains and closest Chinese and American strains (after removal recombinant strains detected by RDP4).