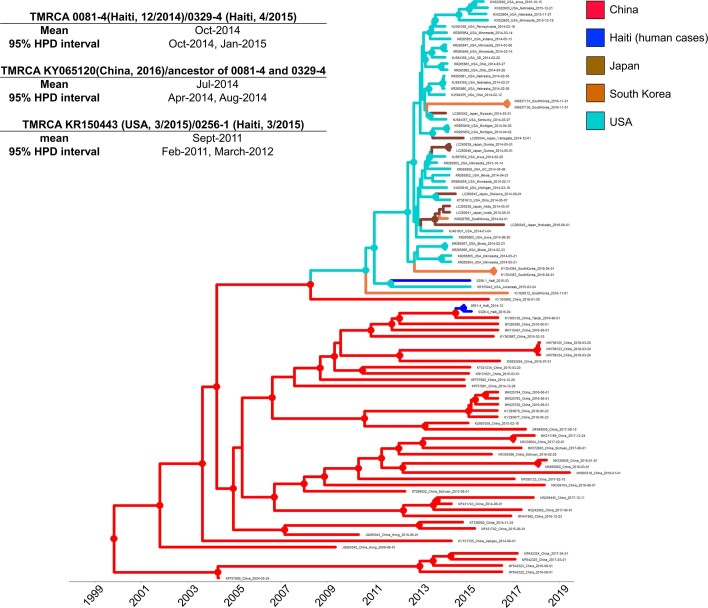
Extended Data Fig. 5. Bayesian maximum clade credibility (MCC) tree of PDCoV strains.
The MCC tree was evolutionary rate. Circles at internal nodes indicate high posterior probability (PP) support >0.9. The table on inferred from a subset of 94 full genome strains that displayed sufficient temporal signal for molecular clock calibration. Branch lengths were scaled in time, according to the bar at the bottom, by using a strict molecular clock and sampling dates to estimate PDCoV the left shows the inferred time of the most recent common ancestor (TMRCA) between Haitian strains and their closest phylogenetic relative, with 95% high posterior density intervals (95%HPD).