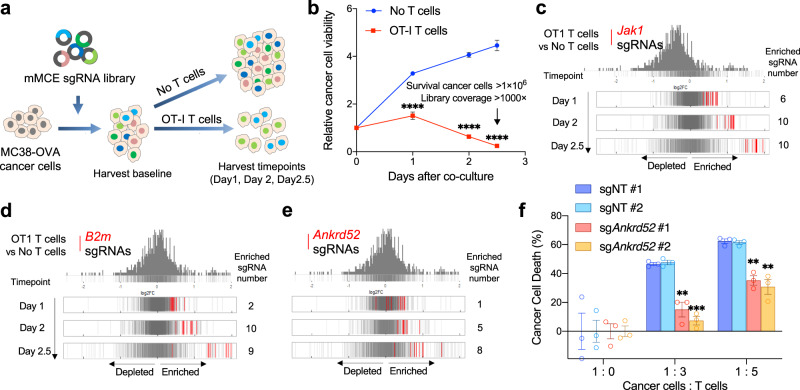
Fig. 3. Targeted in vitro CRISPR screen identifies ANKRD52 as a direct modulator of cancer sensitivity to T cell cytotoxicity.
a Schematic overview of CRISPR screen using co-culture of OT-I T cell with MC38-OVA cells. b Cell viability of MC38-OVA cells co-cultured with OT-I T cells during 2.5 days (n = 3 per group per timepoint). Data are represented as mean ± s.d., ****P < 0.0001, significance is determined using multiple two-tailed Student’s t-test. c–e Distribution histograms of log2FC values for all ten sgRNAs targeting Jak1 (c), B2m (d), or Ankrd52 (e) (Cut-off: FC > 1.4, P < 0.05 for enrichment, analyzed by edgeR). f Killing of MC38-OVA cells with indicated sgRNAs by OT-I T cells at indicated ratio in co-culture. Data are representative of three independent experiments and represented as mean ± s.e.m., **P < 0.01, ***P < 0.001, significance was determined using multiple two-tailed Student’s t-test (sgNT #1 vs sgAnkrd52 #1, P = 3.72×10−3 for 1:3, P = 1.74×10−3 for 1:5; sgNT #2 vs sgAnkrd52 #1, P = 3.64×10−3 for 1:3, P = 1.67×10−3 for 1:5; sgNT #1 vs sgAnkrd52 #2, P = 2.28 × 10−4 for 1:3, P = 4.43 × 10−3 for 1:5; sgNT #2 vs sgAnkrd52 #2, P = 2.7 × 10−4 for 1:3, P = 4.57 × 10−3 for 1:5). See also Supplementary Figs. 6 and 7. Source data are provided as a source data file.