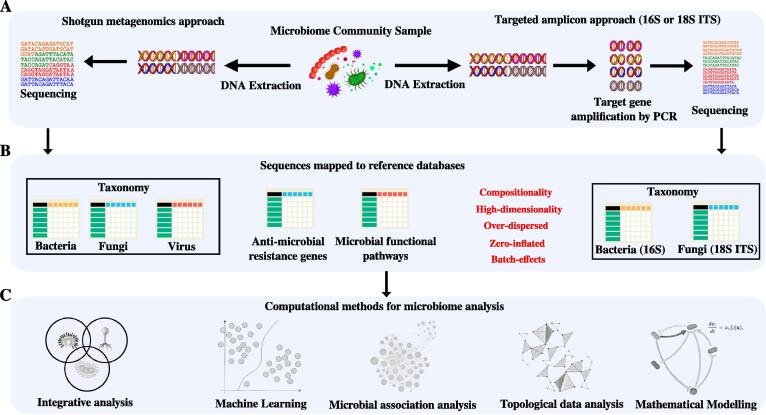
Fig. 1.
Overview of the analytical approaches to microbiome data. (A) Microbiome community samples can be assessed by (1) whole genome shotgun metagenomics: where the whole DNA content is sequenced or (2) Targeted amplicon sequencing: where a targeted region (i.e. 16S in bacteria or ITS in fungi) is amplified by polymerase chain reaction (PCR) followed by sequencing. (B) The derived sequences are next mapped to reference databases to yield taxonomic, anti-microbial resistance or functional profiles of the microbiome (whole genome shotgun metagenomics) or taxonomic profile (targeted amplicon sequencing). Derived microbiome profiles suffer from compositionality, high-dimensionality, over-dispersion, sparsity, and batch effects. (C) Various computational approaches for microbiome analytics can be leveraged including integrative microbiome analysis, machine learning, microbial association analysis, topological data analysis and mathematical modelling.