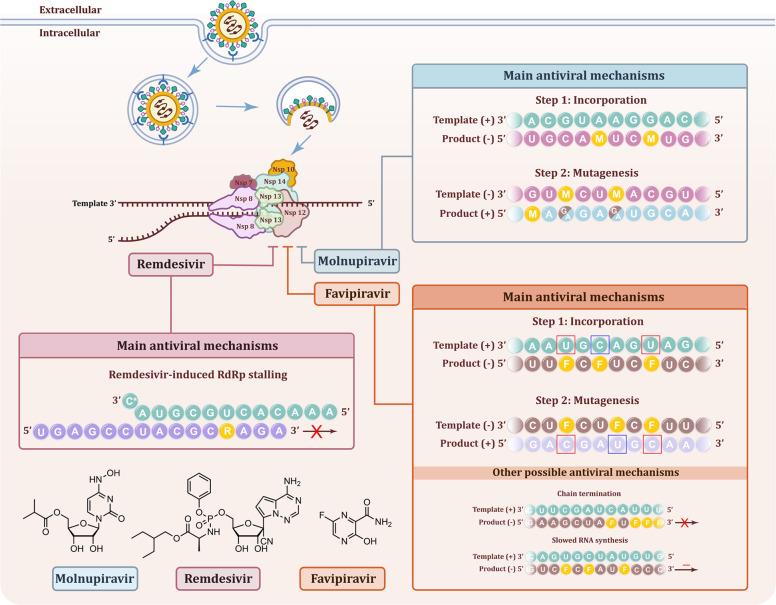
Fig. 1.
Mechanisms of molnupiravir, remdesivir, and favipiravir in against SARS-CoV-2. The virus binds to the ACE2 receptor of the host cell and then be internalized. The membrane of the vacuole fuses with the virus and release the genome of the virus into the cytoplasm of the host cell. The virus genome translated and produced the RNA replicase-transcriptase complex which contains 16 NSPs. One of the NSPs, non-structural protein 12 (Nsp12) has the function of RdRp and could mediate the replication of viral genome together with other replicases. Molnupiravir, remdesivir, and favipiravir could inhibit the function of RdRp. The mechanisms were as follow: (1) Molnupiravir induced RNA mutagenesis by two steps: M nucleotides can be incorporated by RdRp into the negative-strand genomic (−) during copying of the positive-strand template (+) instead of C or U. The obtained negative-strand RNAs containing M can then be used as a template to produce mutagenized positive-strand. These RNA products are predicted to be mutated and results in nonfunctional viruses formated. RNA of random sequence is shown, with M and mutated residues indicated as yellow and brown letters, respectively. (2) Remdesivir acts as a nucleoside analog and could be incorporated into the growing RNA strand by the RdRp. The replacing of ATP with RTP leads to an elongation barrier after addition of three more nucleotides and induces RdRp stalling. RNA of random sequence is shown, with R indicated as yellow. (3) Favipiravir, a purine nucleic acid analog, could mimics the purines G and A to form non-canonical base pairs and has also been found to act against coronaviruses via other mechanisms, including non-obligate chain termination and slowing of RNA synthesis. RNA of random sequence is shown, with F indicated as yellow. The mutated residues are marked with blue or red boxes