Figure 1.
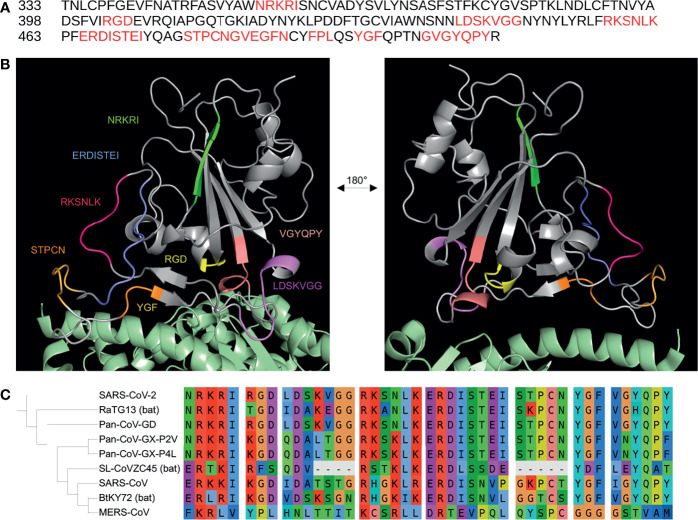
Putative integrin-binding motifs on the SARS-CoV-2 receptor-binding domain.The SARS-CoV-2 receptor-binding domain (RBD) amino acid sequence is shown with the putative integrin-binding motifs marked in red (A). The SARS-CoV-2 spike RBD is depicted with the predicted structurally-accessible integrin-binding motifs highlighted in different colors (B). The spike RBD is shown aligned with the PDB: 6m0j structure in order to show the potential accessibility of these motifs when the spike protein is bound to the canonical host cell entry receptor ACE2 (green) (B). A sequence alignment of the putative integrin-binding motifs found on the SARS-CoV-2 receptor binding domain with spike sequences from the SARS-CoV, RatG13, MERS-CoV, Pan-CoV-GD, Pan-CoV-GX-P4L, Pan-CoV-GX-P2V, SL-CoVZC45, BtKY72 betacoronavirus strains is shown with a corresponding phylogenetic tree of the coronavirus genomes generated using FastTree 2.1 and visualized with iTOL (C).