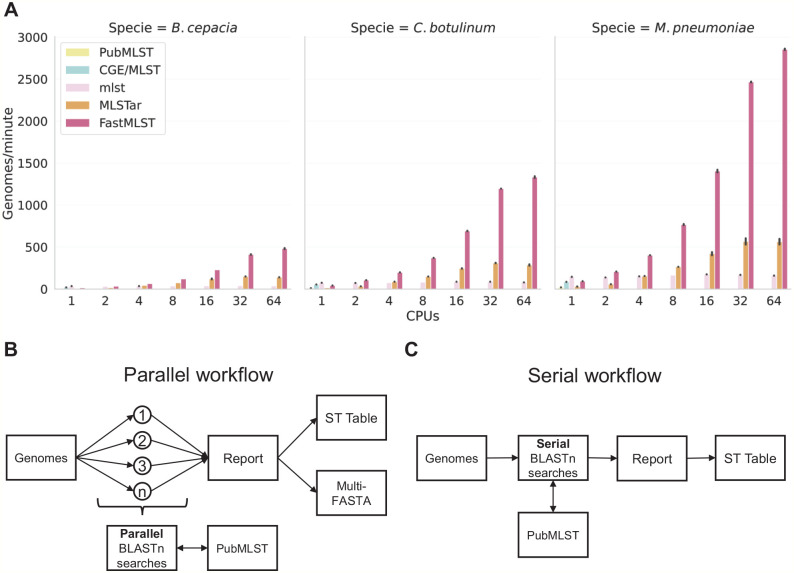
Figure 1.
Processing speed and simplified comparative-workflow. (A) Processing speed (genomes/minute) of FastMLST and standard software for MLST determination (mlst, PubMLST, CGE/MLST, and MLSTar) was compared by triplicate using 3 Burkholderia cepacia (n = 186), Clostridium botulinum (n = 250), and Mycoplasma pneumoniae (n = 175) datasets and 1, 2, 4, 8, 16, 32, and 64 CPUs. Error bars represent the standard error of 3 replicates. Only mlst, MLSTar, and FastMLST natively support parallel processing. (B) FastMLST and MLSTar workflow. (C) mlst, PubMLST, and CGE/MLST workflow. CPU indicates central processing unit; MLST, Multilocus Sequence Typing.