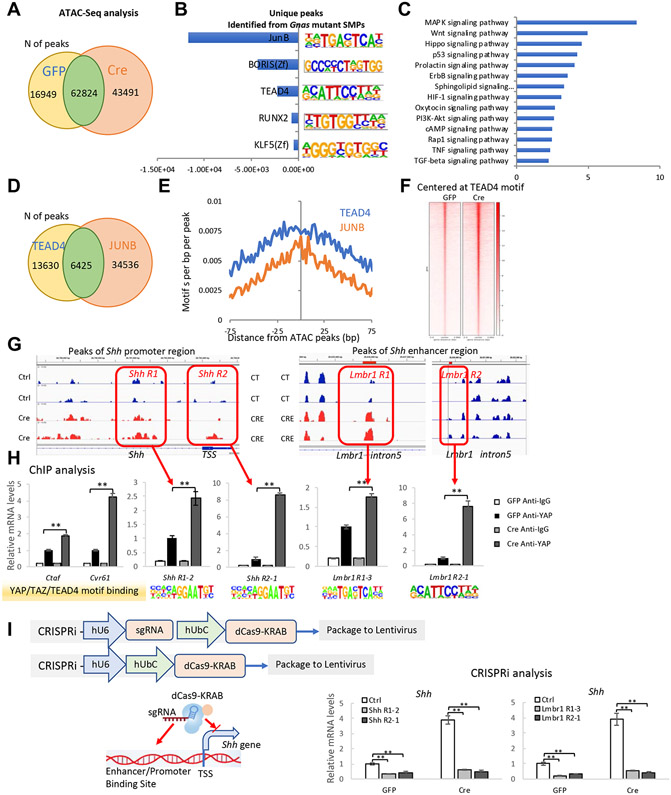
Fig. 5. Shh is a direct target of the YAP transcription factor.
(A) The number of ATAC-seq peaks detected in Ad-GFP- or Ad-Cre-infected Gnasf/f SMPs. (B) HOMER software for unbiased de novo motif discovery was applied to the 43491 unique peaks identified in the Ad-Cre infected cells and the top 5 enriched transcription factor (TF) binding motifs were identified. (C) KEGG-pathway mapping of genes with transcriptional start sites present in the unique ATAC peaks of the Ad-Cre infected Gnasf/f SMPs. (D) The number of JUNB and TEAD4 binding peaks in Ad-Cre infected Gnasf/f SMP cells. (E) The co-occurrence of JunB and TEAD4 binding peaks in the unique ATAC peaks of Ad-Cre-infected Gnasf/f SMPs. (F) ATAC seq signal intensities near TEAD4 motifs. (G) ATAC peaks at TSS and enhancer regions of the SHH locus. (H) ChIP-qPCR analysis of TEAD4 binding sites in each indicated region (boxed). (mean±SD; N=3 biological replicates.). **p<0.01 one-way ANOVA followed by Tukey’s multiple comparisons tests. (I) Shh expression in Ad-GFP or Ad-Cre-infected Gnasf/f SMP cells. TEAD4 sites in the Shh promotor/enhancer regions were occupied by dCas9-KRAB. Schematic is shown in the left panel. QRT-PCR results are shown as mean±SD; N=3 biological replicates. **p<0.01 one-way ANOVA followed by Tukey’s multiple comparisons tests.