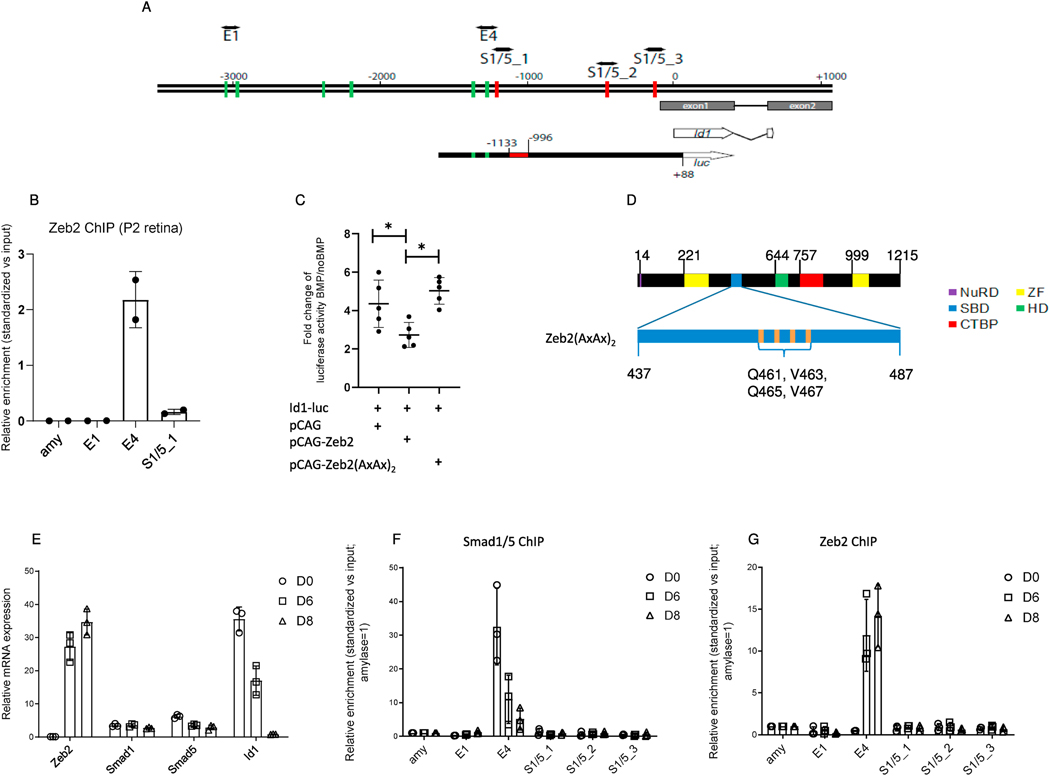
Fig. 5.
BMP-mediated activation of the Id1 promoter. (A) Schematic representation of the mouse Id1 promoter (−3500/+1000, ATG = 0) and comparison with the sequence cloned upstream of the luciferase (luc) gene used for the luciferase assay (Id1-luc). Green lines: Eboxes representing putative Zeb2-binding sites; red lines: putative BMP–Smad-binding elements (indicated as S1/5_1 to S1/5_3). The red box in Id1-luc is the BMP-responsive regulatory region (−1133/−996) in the Id1 promoter. (B) Zeb2 chromatin immunoprecipitation (ChIP) in P2 retinas (N = 2). The enrichment relative to input of the putative Zeb2- (E4, E1) or Smad-binding region (S1/5_1) quantified by PCR is indicated. amy, amylase (C) Scatter plot indicating mean and median fold-change (N = 5) of luciferase activity units of Id1 reporter described in BMP-treated N2a cells expressing either empty vector, wild-type Zeb2 or Zeb2(AxAx)2, compared to non-BMP-treated cells. Significance for differences between the groups was determined by one-way repeated measures ANOVA (F = 11.52, P = 0.0044). Tukey post-hoc test revealed significant differences in BMP-mediated activation between the empty and wild-type Zeb2 groups (P = 0.026), and the wild-type Zeb2 and Zeb2(AxAx)2 groups (P = 0.0041). *P < 0.05. (D) Protein location of the four point mutations in the Smadbinding domain of Zeb2(AxAx)2. (E) mRNA levels of Zeb2, Smad1, Smad5 and Id1 in differentiating murine embryonic stem cells (mESCs). (F) Smad1/5 ChIP and (G) Zeb2 ChIP in neural differentiated mESCs. Putative BMP–Smads::Smad4-binding regions are indicated as S1/5_1 to S1/5_3, while E-boxes are indicated as E1 and E4. BMP–Smads and Zeb2 are recruited in region E4 (−1287/−1147). (F, G) ChIP values are normalized against the input and using amylase (amy) as a negative control (set to 1).