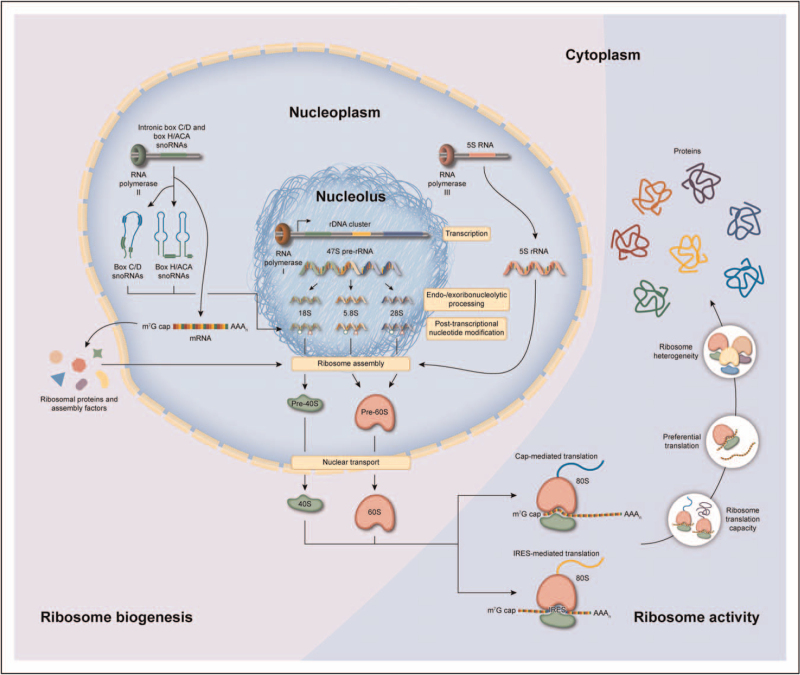
FIGURE 1.
An overview of ribosome biogenesis and activity. 47S prerRNA is transcribed in the nucleolus from rDNA clusters by RNA polymerase I. The 47S precursor is simultaneously processed by endoribonucleases and exoribonucleases and posttranscriptionally modified by snoRNPs. These snoRNPs consist of an enzyme, accessory proteins and box C/D or box H/ACA snoRNAs that guide site-specific 2’-O ribose methylation or pseudouridylation of rRNA nucleotides. These canonical snoRNAs originate from intronic regions of RNA polymerase II-transcribed mRNAs and are liberated by the splicing machinery and subsequent processing. Together with mature rRNA, ribosomal proteins assemble into the small 40S and large 60S ribosomal subunits. This highly coordinated process requires additional assembly factors. After nuclear export, the 40S subunit can form a 43S preinitiation complex together with eukaryotic translation initiation factors that recognize the m7G cap of mRNAs and initiate protein translation after recruitment of the 60S large subunit, resulting in the formation of an active 80S ribosome. In addition to cap-mediated translation, Internal Ribosome Entry Sites (IRESs) can mediate direct recruitment of the ribosome to a translation start site. This process is of paramount importance under cellular stress conditions, where cap-mediated translation is generally inhibited. Ribosome activity is tightly regulated by a multitude of signalling pathways (e.g. AKT/mTOR, TGF/BMP, FGF), and other important factors are energy status and amino acid availability for aminoacyl-tRNA formation. The most rate-limiting step of ribosome activity is translation initiation by eIF4E, which is counteracted by 4E-BP1. In addition, ribosomes can preferentially translate a certain mRNA because of specific (IRES) trans-acting factors. To add to the complexity, the ribosome can generate multiple protein variants from a single mRNA, when more than one or alternative translation initiation sites are present (e.g. in VEGF, MYC and FGF2 mRNAs). Ribosome core protein composition and rRNA posttranscriptional modification levels can vary [4,10], which leads to heterogeneous ribosomes with distinct translational characteristics.