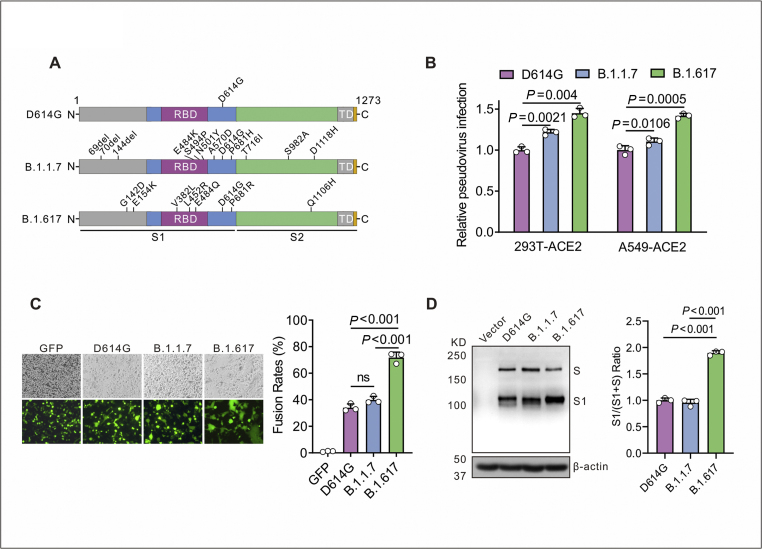
Figure 1.
B.1.617 variant Spike protein of SARS-CoV-2 drives efficient viral entry and cell–cell fusion. (A) The diagram of SARS-CoV-2 Spike protein from D614G, B.1.1.7 and B.1.617 variants. D614G variant pseudovirus (containing the D614G mutation in Spike); B.1.1.7 variant pseudovirus (containing the H69/V70 and Y144 deletions and N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H mutations in Spike); B.1.617 variant pseudovirus (containing the G142D, E154K, V382L, L452R, E484Q, D614G, P681R, and Q1106H mutations in Spike). (B) Infectivity of D614G, B.1.1.7 and B.1.617 variants pseudoviruses was assessed in 293T-ACE2 and A549-ACE2 cells. Cells were inoculated with equivalent doses of each pseudotyped virus, and the supernatants were replaced at 6 h post inoculation with fresh culture. Upon 72 h, cells were lysed with passive lysis buffer and the activity of firefly luciferase was analyzed. (C) Quantitative cell–cell fusion assay. HEK293T cells expressing SARS-CoV-2 Spike variants D614G, B.1.1.7 and B.1.617 were mixed with ACE2-expressing target HEK293T cells (ratio 1: 1), and cell–cell fusion was analyzed by measuring the presence of syncytia by fluorescence microscopy. The fusion rate was the ratio of fluorescence fusion areas/total cell areas under the white light measured by Image Pro-Plus, n = 3. (D) Detection of Spike protein expression of D614G, B.1.1.7 and B.1.617 in HEK 293T cells by Western blot using the anti-RBD (receptor-binding domain) monoclonal antibody. To compare the S1 and S ratio, integrated density of S1/(S + S1) was quantitatively analyzed using ImageJ software, n = 3.