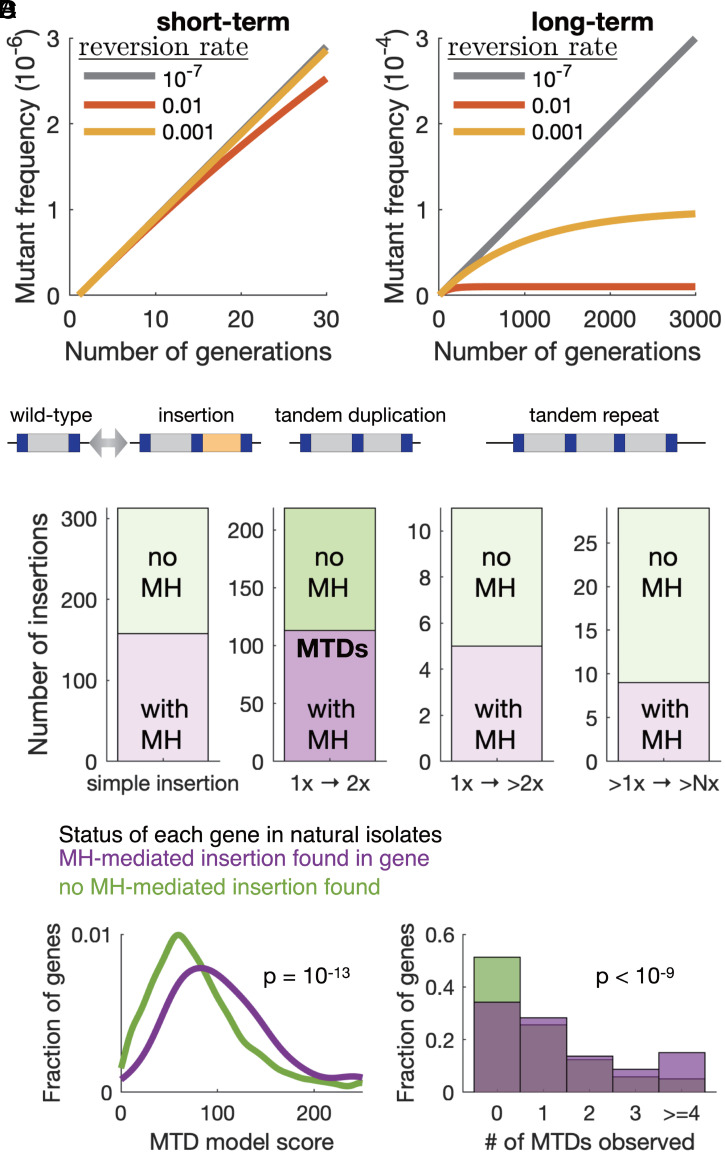
Fig. 5.
MTDs remain subclonal because of high reversion rates, yet half of insertions and de novo tandem duplications in natural populations arise at microhomology sequence pairs. (A) Simulations showing the frequency of a neutral mutation (forward mutation rate = 10−7) within a growing population at three different reversion rates (colors). Left and Right show the same simulates at different timescales, with the effect of reversion only apparent at long timescales. (B) A cartoon showing three possible types of microhomlogy-mediated insertions: simple insertion, tandem duplication, and higher copy repeat. (C) Quantification of all insertions of at least 10 bp fixed in any of the 57 natural S. pombe isolates that represent most of the genetic diversity within the species, relative to the reference genome. Insertions were classified according to the presence (purple) or absence (green) of exact MHPs on either side of the insert and to the type of insert. There are 113 MTDs in wild pombe strains (second column). The right-most column (>1× -> >N×) refers to the expansion of repeats present in the reference genome. (D) Distributions of the predicted MTD score from the logistic regression model (Left) and the number of experimentally observed subclonal MTDs (Right) for genes with one or more microhomology-mediated insertions (purple) or for genes with no MH-mediated insertions (green) in any of the natural isolates. P values are from a Mann–Whitney U test.