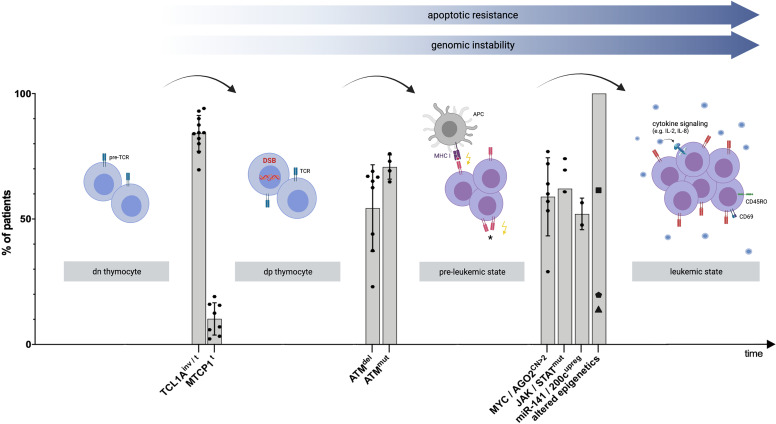
Figure 1.
Proposed model of clonal evolution of T-PLL cells. Schematic visualization explaining T-PLL’s leukemogenesis, based on recent genomic profiling series and corresponding functional assessments. Timeline: Chronology of genomic events leading to the progression to an advanced state of (pre)malignant T-cell development. Y-Axis: Percentage of all analyzed T-PLL patients presenting the respective genomic aberration. Each dot represents a prevalence, derived from selected publications ( Supplementary Table 1 ). The median, as well as standard deviation, out of these publications was calculated for each genomic event. The variability between the studies can be attributed to the different methods and cohort sizes (for more information refer to Supplementary Table 1 ). The first ‘stage’ involves the double-negative (dn) thymocyte, carrying the pre-T cell receptor (pre-TCR) complex. Translocations (t) and inversions (inv) of chromosome 14q at the dp thymocyte stage result in constitutive expression of the proto-oncogenes TCL1A or MTCP1 in a vast majority of T-PLL cases (9, 17). These hits impair the genomic stability of the affected T-cell by reduced DNA repair capacities of DNA double-stranded breaks (DSB) or other (oxidative) insults (10). Deletions (del) and mutations (mut) involving ATM lead to a functionally hypomorphic apical regulator of repair, cell fate, and cell cycle control of the T-PLL precursor. This pre-leukemic cell becomes unable to execute such safeguarding mechanisms upon genotoxic stress (10). Among subsequent perturbations, TCL1A overexpression lowers TCR-signaling thresholds (18), enabling the cell to sustain on low-level input, either by major histocompatibility complex (MHC)-dependent (auto) antigen-presenting cells (APC), or by self-MHC drive only, or by autonomous TCR activation (*, not proven). A central distal node is the JAK/STAT transcriptional machinery. Besides major growth pathways such as the TCR and cytokine-mediated cascades feeding into it, there also is a high prevalence of hyperactivating mutations that target JAK1, JAK3, or STAT5B ( 18) and a high incidence of losses of JAK/STAT negative regulators (43). Further leukemic outgrowth and progression to an exponentially proliferating T-PLL cell are likely mediated by additional aberrations, including copy number (CN) gains on chromosome 8q, leading to MYC amplification and AGO2 overexpression (10). Furthermore, deregulations of T-PLL’s miR-ome, exemplarily represented by the upregulation (upreg) of the miR-141/200c family (45), and of T-PLL’s epigenome in virtually all patients as shown by altered chromatin states at promoters and active enhancers (46), potentially mediated by frequent mutations in KMTs(▪), TET2 (⬟), and EZH2 (▲) (10), contribute to the final leukemic outgrowth of a transformed and activated T-cell (as shown by the T-cell activation marker CD69) with memory-type effector functions (as shown by CD45RO surface expression) (18). The figure was created by the authors using Biorender.com.