Extended Data Figure 5 |. Ligand binding and receptor dimerization are coupled to Pole rotation and PXL changes.
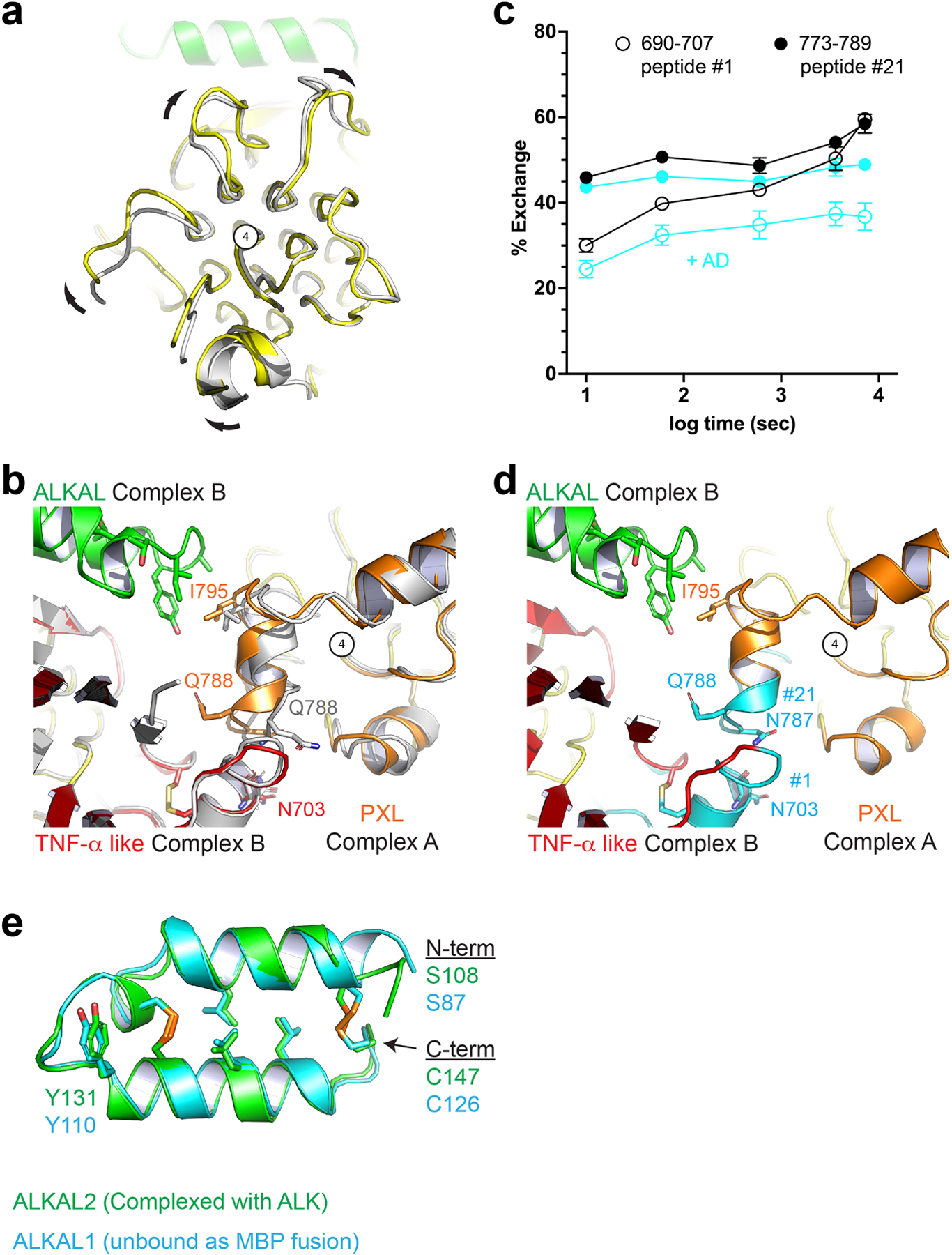
a, View looking down the long axis of the Pole with apo ALK (gray) aligned to the complex structure (color). Compared to unliganded ALK, the ligand-bound ALK dimer undergoes a clockwise rotation of the Pole about the central glycine helix (number 4). b, Unliganded ALK (gray) aligned to both protomers of the complex dimer (color). The PXL residues surrounding Q788 adopt a helical structure upon ligand binding and dimerization. c, Upon ligand binding, two additional peptides – not directly involved in ligand binding and discussed in Fig. 2 – are significantly protected. Statistics were derived from two independent biological repeats, each with three technical repeats. Data represent mean ± SD. d, these peptides (cyan) (peptide #1 and #21, Extended data Fig. 3) correspond to the dimer interface including the disulfide linked helix of the PXL (#21) and the helix on the TNF-α like domain it forms a bond with (#1). e, The core helix-turn-helix of the AD is largely unaltered upon binding. ALKAL2 from the ALK-ALKAL fusion complex structure (green) is aligned to an un-complexed ALKAL1-AD MBP (Maltose Binding Protein) fusion (cyan).
