Abstract
Phytopathogenic fungi have attracted great attention as a promising source for new drug discovery. In the progress of our ongoing study for bioactive natural products from an in-house phytopathogenic fungi library, a pathogenic fungus, Fusarium proliferatum strain 13294 (FP13294), was selected for chemical investigation. Two novel aliphatic unsaturated alcohols named fusariumnols A and B (1 and 2), together with one previously characterized sesquiterpenoid lignoren (3) were identified. Structures of 1–3 were assigned by mass spectrometry and NMR spectroscopy. Their bioactivities were assessed against Staphylococcus epidermidis, S. aureus, and Methicillin-resistant S. aureus (MRSA). Compounds 1 and 2 exhibited weak antibacterial activity against S. epidermidis (MIC = 100 μM).
Keywords: Pathogenic fungus, Aliphatic unsaturated alcohols, Antibacterial, Natural products
1. Introduction
Fungi inhabiting special environments are well-known producers of secondary metabolites with diverse biological activities. Phytopathogenic fungi have attracted increasing attention because of frequent discoveries of natural products with diverse structural properties and promising biological activities [1,2]. These are treasure troves for mining novel products especially in phytopathogenic fungi where genes have arisen during the long-term co-evolutionary process with host plants; these gene products have high potential in pharmaceutical and agricultural applications [3,4]. For example, phytotoxins derived from phytopathogenic fungi have been used as ecofriendly tools in the development of safe bioherbicides [5,6]. In addition, fungal phytotoxins exhibited a variety of biological activities, including promising antifungal, anticancer, and anti-inflammatory activities [[7], [8], [9], [10]].
The genus Fusarium, often isolated from different plant tissues and from plant debris, can afford diverse mycotoxins, causing reductions of crop quality and harvest, as well as effecting human health [11]. Nevertheless, there is abundant evidence indicating Fusarium sp. have the capacity to afford different kinds of natural products, such as polyketides, terpenoids, alkaloids, and peptides, which show significant biological activities [[12], [13], [14], [15]]. F. proliferatum is a widespread plant pathogenic fungi associated with diverse crops, including rice, wheat, maize, garlic, asparagus, date palm, and Chinese chive [16]. F. proliferatum has been reported to produce multiple mycotoxins, such as fumonisins, beauvericin, fusaproliferin, moniliformin, and fusaric acid [17].
Staphylococci, which are Gram-positive bacteria, can colonize humans and cause infections as the immune system weakens. Pathogens are introduced through wounds and also associated with the insertion of medical devices [18,19]. They are able to form biofilms in chronic wounds, resulting in doubling of time for recovery. Due to extremely difficult to treat, these infections lead to a serious burden for the public health system [20]. The Staphylococcus species usually contain S. aureus and S. epidermidis. Because of increased bacterial resistance to current antibiotics in clinical use, there is a need to explore natural products to identify novel compounds for drug discovery.
During an ongoing study of novel bioactive natural products in our in-house phytopathogenic fungi library, a strain of F. proliferatum (FP13294) was subjected to investigate secondary metabolites based on bioassay guided strategy. Its crude extract showed weak antibacterial activity against S. epidermidis (MIC = 200 μM). Fractionation of crude extracts obtained from a rice fermentation culture yielded two new secondary metabolites, which we named fusariumnols A and B (1 and 2), in addition to one known compound lignoren (3). We herein provide details of the structure identification and biological activities of compounds 1–3.
2. Materials and methods
2.1. General experimental procedures
1D and 2D NMR spectra were measured on a Bruker Avance DRX 600 MHz spectrometer with TCI cryoprobes. Chemical shifts are expressed as δ (ppm) referenced to the solvent peaks at δC 49.0 and δH 3.31 for Methanol-d4. HRESIMS spectra were measured on a Thermo Orbitrap Q Exactive mass spectrometer. Materials including Sephadex LH-20 (GE Healthcare) and ODS-A (YMC) were utilized for column chromatography. Reverse Phase HPLC chromatography was carried out by an Agilent 1100 Series HPLC, and ChemStation Rev.B.02.01 software was used to analysis the data. Semipreparative RP-HPLC was performed equipped with an ACE Excel 5C18-AR column (10 × 250 mm, 5 μm).
2.2. Microbial strain culture and identification of F. proliferatum 13,294
F. proliferatum 13294 (FP13294) was prepared on potato dextrose agar (PDA) at 28 °C. Strain FP13294 was identified based on morphology and 18S rRNA gene sequence. The primer pair EF-1a-F (5″-AAGGCTGGTTCCAAGACTGG-3″) and EF-1a-R (5″-TGGTCGTCTCTTTCGCTCCT-3″) were used to amplify the translation elongation factor 1 alpha (EF-1a) gene region of FP13294. The PCR amplification (25 μL in total) contained 1 μL of DNA template, 0.4 μL of rTaq polymerase, 0.4 μL of each primer, 2.5 μL of 2.5 nM dNTP, and 2.5 μL of 10 × buffer. The PCR reaction was carried out on an ABI PCR Thermal Cycler with following condition: denaturation at 94 °C for 5 min, 25 cycles of denaturation at 94 °C for1 min, annealing at 55 °C for 1 min, elongation at 72 °C for 45 s, and a final elongation at 72 °C for 10 min. Both forward and reverse primers were used for PCR product sequencing, and the consensus sequence was compared to the GenBank database using BLASTn. Sequences with high similarity were acquired from GenBank. Phylogenetic neighbor-joining tree was constructed by MEGA 6.0 software [21], and rooted on Trichoderma auranteffusum.
2.3. Fermentation and isolation of compounds1–3
FP13294 was prepared on PDA plate medium at 28 °C for 7 days, and agar plugs (0.5 × 0.5 × 0.5 cm3) were inoculated into six Erlenmeyer flasks (250 mL) each containing 100 mL potato dextrose broth (PDB). Then the Erlenmeyer flasks were incubated for five days at 28 °C on a rotary shaker at 200 rpm as the seed culture. Fermentation was then carried out in 200 aseptic-bags each containing 80 g of autoclaved rice and 120 mL sterilized distilled H2O for 30 days at 28 °C. The fermentation material was extracted with EtOAc three times, and then was evaporated under in vacuo to give an extract (18.64 g).
The crude extract was subjected to Sephadex LH-20 eluting with 100% MeOH to yield seven fractions (A–G). Fraction E (11.33 g) was fractionated by Sephadex LH-20 again eluting with100% MeOH to give four sub-fractions (E1–E4). Then Fraction E3 (7.34 g) was subjected to ODS-MPLC with an ACN-H2O gradient (5%–100%) elution for 100 min to get ten sub-fractions (E3A–E3J). Sub-fraction E3C (2.56 g) was further purified by Sephadex LH-20 eluting with 100% MeOH to afford seven fractions (E3C1–E3C16). Fraction E3C4 (641 mg) was chromatographed over ODS-MPLC with an ACN-H2O gradient (10%–100%) elution for 100 min to yield eight sub-fractions (E3C4A–E3C4H). Subfraction E3C4D (148.2 mg) was then purified by semipreparative RP-HPLC equipped with an ACE Excel 5C18-AR (10 × 250 mm) eluting with 35% ACN-H2O for 60 min (flow rate: 4 ml/min) to obtain 1 (2.4 mg, tR = 30.9 min), 2 (0.5 mg, tR = 52.9 min), and 3 (17.4 mg, tR = 21.4 min).
2.4. Accession number
Fusarium proliferatum stain 13,294 has been deposited at the China General Microbiological Culture Collection Center with the accession number 21945. The sequence data of the EF-1a gene has been deposited under GenBank accession number MZ382914.
3. Results
3.1. Characterization and identification of strain FP13294
Isolate 13294 was originally obtained from wheat tissue. It appeared as white villous colonies which produce light purple pigment and have a colony diameter of 7 mm after 7 d on potato dextrose agar (PDA) at 28 °C (Fig. 1). The EF-1a gene sequence and neighbor-joining tree (Fig. 1) confirmed it as Fusarium proliferatum.
Fig. 1.
Morphology and phylogenetic tree of F. proliferatum 13294. a Morphology characteristic of FP13294 cultured on PDA at 28 °C for 7 days. b Phylogenetic neighbor-joining tree of FP13294 based on EF-1a gene sequences. NCBI accession numbers are given in parentheses. Numbers at nodes indicate levels of bootstrap support (percentage) based on 1000 resampled datasets; only values > 50% are presented. The bar indicates 0.5 amino acid substitutions per site. Trichoderma auranteffusum was chosen as outgroup.
3.2. Structure elucidation
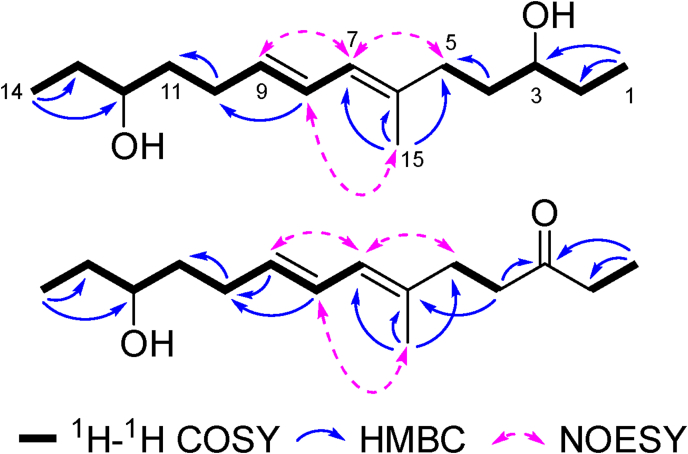
The HRESIMS (Fig. S1a) analysis revealed the molecular formula of 1 as C15H28O2 (241.2162 [M + H]+, calcd. for 241.2162), implying two degrees of unsaturation. Based on combined analysis of the 1H, 13C, and HSQC spectra of 1 (Table 1 and Figs. S1b, S1c, S1e), 15 carbon resonances could be observed, composed of one olefinic quaternary carbon (δC 137.1), three conjugated sp2 methines [δC/δH 133.0/5.57 (dt, J = 15.2, 6.9 Hz), 128.3/6.29 (dd, J = 15.2, 10.5 Hz), 126.3/5.83 (d, J = 10.5 Hz)], two oxymethine groups [δC/δH 73.7/3.44 (m), 73.4/3.47 (m)], six sp3 methylenes, two terminal methyl triplets [2 × δC/δH 10.5/0.95 (t, J = 7.4 Hz)], and one methyl singlet [δC/δH 16.7/1.72 (s)], indicating that the structure of 1 comprises a bishydroxylated alkyl chain with two conjugated olefinic groups. The 1H–1H COSY (Fig. S1d) correlations of 1 revealed the existence of two sub-units C-1–C-5 and C-7–C-14, which were deduced to connect at C-6 based on the key HMBC (Fig. S1f) correlations of H3-15 with C-5/C-6/C-7. Further HMBC analysis revealed that the two hydroxyl groups were substituted at C-3 and C-12, based on the crosspeaks between H3-1/C-3 and H3-14/C-12. The NOESY (Fig. S1g) correlations between H3-15/H-8, H-5/H-7, and H-7/H-9 together with a large coupling constant between H-8/H-9 (3JH-H = 15.2 Hz) assigned the Δ6,7 and Δ8,9 double bonds as E configuration. To determine the relative configurations between C-3 and C-12, the density functional theory (DFT) based 13C NMR calculation and DP4 analysis of two epimers 1a/1b was conducted, resulting in higher Bayes's theorem probability of 1b (75%) compared with 1a (25%) (Table S1). Therefore, the structure of 1 was established as illustrated in Fig. 3 and named fusariumnol A.
Table 1.
1H and13C NMR Data of compounds 1 and 2.
| Pos. | 1 |
2 |
||
|---|---|---|---|---|
| δHa mult (J in Hz) | δCb, type | δHa mult (J in Hz) | δCb, type | |
| 1 | 0.95, t (7.4) | 10.5, CH3 | 1.00, t (7.4) | 8.0, CH3 |
| 2 | 1.44, m | 31.2, CH2 | 2.47, q (7.3) | 35.1, CH2 |
| 3 | 3.44, m | 73.7, CH | 214.0, C | |
| 4a | 1.59, m | 36.4, CH2 | 2.56, t (7.6) | 40.2, CH2 |
| 4b | 1.51, m | |||
| 5a | 2.19, m | 37.1, CH2 | 2.27, t (7.6) | 34.8, CH2 |
| 5b | 2.08, m | |||
| 6 | 137.1, C | 135.6, C | ||
| 7 | 5.83, d (10.5) | 126.3, CH | 5.77, d (10.7) | 126.5, CH |
| 8 | 6.29, dd (15.2, 10.7) | 128.3, CH | 6.25, dd (15.2, 10.7) | 128.0, CH |
| 9 | 5.57, dt (15.1, 6.9) | 133.0, CH | 5.56, dt (15.1, 7.1) | 133.5, CH |
| 10a | 2.24, m | 30.3, CH2 | 2.21, m | 30.1, CH2 |
| 10b | 2.15, m | 2.13, m | ||
| 11a | 1.54, m | 38.0, CH2 | 1.54, m | 37.8, CH2 |
| 11b | 1.49, m | 1.49, m | ||
| 12 | 3.47, m | 73.4, CH | 3.44, m | 73.3, CH |
| 13a | 1.51, m | 31.2, CH2 | 1.48, m | 31.1, CH2 |
| 13b | 1.41, m | |||
| 14 | 0.95, t (7.4) | 10.5, CH3 | 0.93, t (7.5) | 10.3, CH3 |
| 15 | 1.72, s | 16.7, CH3 | 1.75, s | 16.7, CH3 |
Recorded at 600 MHz in Methanol-d4.
Recorded at 125 MHz in Methanol-d4.
Fig. 3.
Key 2D NMR correlations of compounds 1 and 2.
The molecular formula of 2 could be deduced as C15H26O2 with three degrees of unsaturation from its HRESIMS (Fig. S2a) spectrum (239.2005 [M + H]+, calcd. for 239.2006). The 1H and 13C NMR data of 2 (Figs. S2b and S2c) showed high similarity with those of 1, except for an additional keto-carbonyl group (δC 214.0) replacing one of the original O-substituted methines in 1. Further HMBC (Fig. S2f) analysis revealed that compound 2 is the 3-oxo derivative of 1 based on crosspeaks of H3-1 with C-2/C-3, and H2-4 with C-3. The Δ6,7 and Δ8,9 double bonds were also determined to be E configuration based on the NOESY correlations (Fig. S2g) between H3-15/H-8, H-5/H-7, and H-7/H-9 combined with 3JH8-H9 value (15.1 Hz). Compound 2 was proposed as a new analog of 1 and was named fusariumnol B (Fig. 2).
Fig. 2.
Structure of isolated compounds (1–3) from F. proliferatum 13294.
The structure of one known compound, lignoren (3), was determined based on the HRESIMS, 1H and 13C NMR data (Figs. S3a–S3c) and by comparison with those of published work [22].
3.3. Bioactivity tests
The crude extract, purified subfractions, and isolated compounds 1–3 were assessed for their anti-bacterial activity against Gram-positive bacterium, including S. aureus, MRSA, and S. epidermidis. The crude extract and subfraction E3C4D showed anti-bacterial activity against S. epidermidis (MIC = 200 μM). Finally, we found that compounds 1 and 2 exhibited weak anti-bacterial activity against S. epidermidis (MIC = 100 μM).
4. Discussion and conclusion
The discovery of penicillin initiated an era wherein natural products derived from microorganisms revolutionized medicine. Afterwards, human life expectancy was extended by nearly 40 years [23]. Historically, the majority of approved drugs have been found from natural sources or derivatives of natural compounds. However, due to the repeat discovery of known compounds in other microorganisms, the discovery ratio of new natural compounds with bioactivity has declined in recent years. Additionally, many pharmaceutical companies have reduced their investment in natural product research. Thus, to further advance drug discovery, new strategies for the discovery of novel natural products are of great importance.
Recently, poorly tapped biological resources, including rare actinobacteria, endophytic fungi, and even phytopathogenic fungi, have been explored as innovative resources for discovering novel natural products with promising bioactivity [[24], [25], [26]]. Among them, phytopathogenic fungi play an important, but yet rarely explored role. They are classified as biotrophs, hemibiotrophs, and necrotrophs, acquiring nutrition during invading process of their host plants. Between the interactions of phytopathogenic fungi and plants, fungi usually metabolize and produce series of low-molecular weight secondary metabolites, which are not essential for life of phytopathogenic fungi while are known for their versatility. These secondary metabolites always play an important role in ecological defense and biological competition. In association with host plants, phytopathogenic fungi can become rich sources of cyclic peptides, terpenes, alkaloids, aliphatic hydrocarbons, aromatic polyketones, and heterocyclic compounds, showing special values to humans [27]. Recently, there has been great progress in the research of the secondary metabolites from phytopathogenic fungi. Many natural products they produce e.g., sirodesmin PL [28,29], AF-toxin Ⅰ/II [30], and cytochalasins B [31] show different promising bioactivities. Fusarium species are distributed worldwide, and mainly attack grain crops causing quality and yield reductions; they also produce a variety of mycotoxins, such as corn gibberelenone [11], beauvericin [32], fusaproliferin [33], and fumonisin [34]. In addition, abundant active secondary metabolites have also been isolated from Fusarium sp., including antibacterial and anticancer naphthoquinone derivatives [35,36], anticancer and anti-inflammatory alkaloids [37,38], antibacterial terpenoids [39], antifungal and anti-malarial cyclodepsipeptide [40], etc. For example, F. solani has the ability to produce cyclooxygenase-2 (COX-2) inhibitor fusopoltide A [41] and anti-tumor naphthoquinone compound solaninaphthoquione [42]. Kakeya et al. reported the isolation and structure elucidation of lucilactaene with unique structure, a cell cycle inhibitor derived from Fusarium sp. RK 97-94 [43]. Renner et al. focused on marine Fusarium species to mine novel bioactive secondary metabolites. Neomangicols, halogenated sesterterpenoids with cytotoxic activity, and mangicols A−G, sesterterpene polyols with remarkable anti-inflammatory activity, were identified from F. heterosporum [44]. In order to obtain alternatives of biohazardous nematocides, Bogner et al. got three secondary metabolites, gibepyrone D, indole-3-acetic acid, and 4-hydroxybenzoic acid, with prominent nematode-antagonistic activity from strain F. oxysporum 162 [45]. These results provided clues for the finding of novel biohazardous nematocides. Parnafungin A and B were initially isolated from F. larvarum, as antifungal agents. In addition, antiviral medicine podophyllotoxin [46], anti-malaria drug quinine and cinchonidine [47], as well as cardiovascular agent ginkgolide B [48], were reported to be prepared from Fusarium species. Furthermore, F. proliferatum is known to produce a variety of mycotoxins [49,50]. These mycotoxins can increase health risks when fungal contaminated agricultural products are consumed. However, there are many secondary metabolites from these species which have not been investigated.
This study on the pathogen F. proliferatum isolate FP13294 expands the knowledge about the chemical production and biological capabilities of the genus Fusarium. Compounds 1–2 were reported here for the first time and belongs to the alkenol family. Compound 3 is a sesquiterpenoid, identified from Trichoderma lignorum in 2004 [22] but have not yet been reported in the genus Fusarium. Our results showed compounds 1 and 2 exhibited weak antibacterial activity against S. epidermidi (MIC = 100 μM).
Alkenols are widely distributed in nature and are reported to be utilized as antibacterial, anti-inflammatory and anti-tumor agents, and even as pheromones. Avocadene was reported to exhibit anti-bacterial as well as anti-inflammatory activities [51]. As a component of many essential oils, linalool showed anti-tumor effect [52] and was used in leukemia therapy [53]. In addition, 11-eicosen-1-ol was found to be the major ingredient of alarm pheromones of worker honeybee [54]. Therefore, potentiality of alkenol no matter in bioactive drugs development than in interaction progress between pathogen and host plants are cause for concern.
CRediT authorship contribution statement
Wanying Lu: Visualization, Investigation, Writing – original draft. Guoliang Zhu: Visualization, Investigation, Writing – original draft. Weize Yuan: Investigation. Zhaoxi Han: Investigation. Huanqin Dai: Investigation, Activity screening. Mostafa Basiony: Writing – review & editing. Lixin Zhang: Conceptualization, Supervision. Xueting Liu: Conceptualization, Supervision. Tom Hsiang: Resources, Writing – review & editing. Jingyu Zhang: Conceptualization, Supervision, Writing – original draft.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2020YFA0907200, 2019YFA0906200, and 2020YFA0907800), the National Natural Science Foundation of China (21877038, 21907031, 21977029, 31720103901, and 81903529), Shanghai Rising-Star Program (20QA1402800), the Open Project Funding of the State Key Laboratory of Bioreactor Engineering, and the 111 Project (B18022). Discovery and isolation of F. proliferatum strain 13294 was supported by the Natural Sciences and Engineering Research Council of Canada funding to T. Hsiang.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.10.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Blackwell M. The Fungi: 1, 2, 3… 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Stergiopoulos I., Collemare J., Mehrabi R., De Wit P.J. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- 3.Kusari S., Hertweck C., Spiteller M. Chemical ecology of endophytic fungi: origins of secondary metabolites. Chem Biol. 2012;19:792–798. doi: 10.1016/j.chembiol.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J.Y., Wang Z.Z., Song Z.J., Karthik L., Hou C.J., Zhu G.L., et al. Brocaeloid D, a novel compound isolated from a wheat pathogenic fungus, Microdochium majus 99049. Synth Syst Biotechnol. 2019;4:173–179. doi: 10.1016/j.synbio.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbas H.K., Duke S.O. Phytotoxins from plant pathogens as potential herbicides. J Toxicol Toxin Rev. 1995;14:523–543. doi: 10.3109/15569549509016440. [DOI] [Google Scholar]
- 6.Sugawara F., Strobel G., Fisher L., Van Duyne G., Clardy J. Bipolaroxin, a selective phytotoxin produced by Bipolaris cynodontis. Proc Natl Acad Sci U S A. 1985;82:8291–8294. doi: 10.1073/pnas.82.24.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon R.A. Natural products and plant disease resistance. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 8.Hammerschmidt R. Phytoalexins: what have we learned after 60 years? Annu Rev Phytopathol. 1999;37:285–306. doi: 10.1146/annurev.phyto.37.1.285. [DOI] [PubMed] [Google Scholar]
- 9.Li F.L., Sun W.G., Guan J.K., Lu Y.Y., Lin S., Zhang S.T., et al. Anti-inflammatory fusicoccane-type diterpenoids from the phytopathogenic fungus Alternaria brassicicola. Org Biomol Chem. 2018;16:8751–8760. doi: 10.1039/c8ob02353k. [DOI] [PubMed] [Google Scholar]
- 10.Osbourn A.E. Antimicrobial phytoprotectants and fungal pathogens: a commentary. Fungal Genet Biol. 1999;26:163–168. doi: 10.1006/fgbi.1999.1133. [DOI] [PubMed] [Google Scholar]
- 11.Dweba C., Figlan S., Shimelis H., Motaung T., Sydenham S., Mwadzingeni L., et al. Fusarium head blight of wheat: pathogenesis and control strategies. Crop Protect. 2017;91:114–122. doi: 10.1016/j.cropro.2016.10.002. [DOI] [Google Scholar]
- 12.Elavarasi A., Rathna G.S., Kalaiselvam M. Taxol producing mangrove endophytic fungi Fusarium oxysporum from Rhizophora annamalayana. Asian Pac J Trop Biomed. 2012;2:S1081–S1085. doi: 10.1016/S2221-1691(12)60365-7. [DOI] [Google Scholar]
- 13.Jayasinghe L., Abbas H.K., Jacob M.R., Herath W.H., Nanayakkara N.D. N-Methyl-4-hydroxy-2-pyridinone analogues from Fusarium oxysporum. J Nat Prod (Gorakhpur) 2006;69:439–442. doi: 10.1021/np050487v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar A., Patil D., Rajamohanan P.R., Ahmad A. Isolation, purification and characterization of vinblastine and vincristine from endophytic fungus Fusarium oxysporum isolated from Catharanthus roseus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q.X., Li S.F., Zhao F., Dai H.Q., Bao L., Ding R., et al. Chemical constituents from endophytic fungus Fusarium oxysporum. Fitoterapia. 2011;82:777–781. doi: 10.1016/j.fitote.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Proctor R.H., Desjardins A.E., Moretti A. Springer; 2009. Biological and chemical complexity of Fusarium proliferatum. the role of plant pathology in food safety and food security; pp. 97–111. [DOI] [Google Scholar]
- 17.Jennings P. Fusarium mycotoxins: chemistry, genetics and biology‐by Anne E. Desjardins. Plant Pathol. 2007;56:337–338. doi: 10.1111/j.1365-3059.2006.01505.x. [DOI] [Google Scholar]
- 18.Vuong C., Otto M. Staphylococcus epidermidis infections. Microb Infect. 2002;4:481–489. doi: 10.1016/s1286–4579(02)01563-0. [DOI] [PubMed] [Google Scholar]
- 19.Yao Y., Sturdevant D.E., Villaruz A., Xu L., Gao Q., Otto M. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect Immun. 2005;73:1856–1860. doi: 10.1128/iai.73.3.1856-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowling F.L., Jude E.B., Boulton A.J. MRSA and diabetic foot wounds: contaminating or infecting organisms? Curr Diabetes Rep. 2009;9:440–444. doi: 10.1007/s11892-009-0072-z. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg A., Kemami Wangun H.V., Nkengfack A.E., Schlegel B. Lignoren, a new sesquiterpenoid metabolite from Trichoderma lignorum HKI 0257. J Basic Microbiol. 2004;44:317–319. doi: 10.1002/jobm.200410383. [DOI] [PubMed] [Google Scholar]
- 23.Li J.W.-H., Vederas J.C. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 24.Radić N., Štrukelj B. Endophytic fungi—the treasure chest of antibacterial substances. Phytomedicine. 2012;19:1270–1284. doi: 10.1016/j.phymed.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q., Song F.H., Xiao X., Huang P., Li L., Monte A., et al. Abyssomicins from the South China Sea deep‐sea sediment Verrucosispora sp.: natural thioether Michael addition adducts as antitubercular prodrugs. Angew Chem Int Ed Engl. 2013;125:1269–1272. doi: 10.1002/anie.201208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Zhang J.Y., Ren B., Lu W.Y., Hou C.J., Wang J., et al. Characterization of anti-BCG benz [α] anthraquinones and new siderophores from a Xinjiang desert–isolated rare actinomycete Nocardia sp. XJ31. Appl Microbiol Biotechnol. 2020;104:8267–8278. doi: 10.1007/s00253-020-10842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brase S., Encinas A., Keck J., Nising C.F. Chemistry and biology of mycotoxins and related fungal metabolites. Chem Rev. 2009;109:3903–3990. doi: 10.1021/cr050001f. [DOI] [PubMed] [Google Scholar]
- 28.Gardiner D.M., Cozijnsen A.J., Wilson L.M., Pedras M.S.C., Howlett B.J. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol Microbiol. 2004;53:1307–1318. doi: 10.1111/j.1365-2958.2004.04215.x. [DOI] [PubMed] [Google Scholar]
- 29.Rouxel T., Chupeau Y., Fritz R., Kollmann A., Bousquet J.-F. Biological effects of sirodesmin PL, a phytotoxin produced by Leptosphaeria maculans. Plant Sci. 1988;57:45–53. doi: 10.1016/0168-9452(88)90140-9. [DOI] [Google Scholar]
- 30.Ito K., Tanaka T., Hatta R., Yamamoto M., Akimitsu K., Tsuge T. Dissection of the host range of the fungal plant pathogen Alternaria alternata by modification of secondary metabolism. Mol Microbiol. 2004;52:399–411. doi: 10.1111/j.1365-2958.2004.04004.x. [DOI] [PubMed] [Google Scholar]
- 31.Evidente A., Andolfi A., Vurro M., Zonno M.C., Motta A. Cytochalasins Z1, Z2 and Z3, three 24-oxa [14] cytochalasans produced by Pyrenophora semeniperda. Phytochemistry. 2002;60:45–53. doi: 10.1016/S0031-9422(02)00071-7. [DOI] [PubMed] [Google Scholar]
- 32.Moretti A., Logrieco A., Bottalico A., Ritieni A., Randazzo G. Production of beauvericin by Fusarium proliferatum from maize in Italy. Mycotoxin Res. 1994;10:73–78. doi: 10.1007/BF03192255. [DOI] [PubMed] [Google Scholar]
- 33.Santini A., Ritieni A., Fogliano V., Randazzo G., Mannina L., Logrieco A., et al. Structure and absolute stereochemistry of fusaproliferin, a toxic metabolite from Fusarium proliferatum. J Nat Prod. 1996;59:109–112. doi: 10.1021/np960023k. [DOI] [PubMed] [Google Scholar]
- 34.Gelderblom W., Jaskiewicz K., Marasas W., Thiel P., Horak R., Vleggaar R., et al. Fumonisins--novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl Environ Microbiol. 1988;54:1806–1811. doi: 10.1128/aem.54.7.1806-1811.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Supratman U., Hirai N., Sato S., Watanabe K., Malik A., Annas S., et al. New naphthoquinone derivatives from Fusarium napiforme of a mangrove plant. Nat Prod Res. 2021;35:1406–1412. doi: 10.1080/14786419.2019.1650358. [DOI] [PubMed] [Google Scholar]
- 36.Tadpetch K., Chukong C., Jeanmard L., Thiraporn A., Rukachaisirikul V., Phongpaichit S., et al. Cytotoxic naphthoquinone and a new succinate ester from the soil fungus Fusarium solani PSU-RSPG227. Phytochem Lett. 2015;11:106–110. doi: 10.1016/j.phytol.2014.11.018. [DOI] [Google Scholar]
- 37.Chowdhury N.S., Sohrab M.H., Rana M.S., Hasan C.M., Jamshidi S., Rahman K.M. Cytotoxic naphthoquinone and azaanthraquinone derivatives from an endophytic Fusarium solani. J Nat Prod (Gorakhpur) 2017;80:1173–1177. doi: 10.1021/acs.jnatprod.6b00610. [DOI] [PubMed] [Google Scholar]
- 38.Sun W.J., Zhu H.T., Zhang T.Y., Zhang M.Y., Wang D., Yang C.R., et al. Two new alkaloids from Fusarium tricinctum SYPF 7082, an endophyte from the root of Panax notoginseng. Nat Prod Bioprospect. 2018;8:391–396. doi: 10.1007/s13659-018-0171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong J.W., Cai L., Li X.J., Duan R.T., Shu Y., Chen F.Y., et al. Production of a new tetracyclic triterpene sulfate metabolite sambacide by solid-state cultivated Fusarium sambucinum B10.2 using potato as substrate. Bioresour Technol. 2016;218:1266–1270. doi: 10.1016/j.biortech.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 40.Ibrahim S.R., Abdallah H.M., Elkhayat E.S., Al Musayeib N.M., Asfour H.Z., Zayed M.F., et al. Fusaripeptide A: new antifungal and anti-malarial cyclodepsipeptide from the endophytic fungus Fusarium sp. J Asian Nat Prod Res. 2018;20:75–85. doi: 10.1080/10286020.2017.1320989. [DOI] [PubMed] [Google Scholar]
- 41.Chen K., Sun W., Bie Q., Liu X., Chen C., Liu J., et al. Fusopoltide A and fusosterede A, A polyketide with a pentaleno [1, 2-c] pyran ring system and A degraded steride, from the fungus Fusarium solani. Tetrahedron Lett. 2018;59:2679–2682. doi: 10.1016/j.tetlet.2018.05.082. [DOI] [Google Scholar]
- 42.Tadpetch K., Chukong C., Jeanmard L., Thiraporn A., Rukachaisirikul V., Phongpaichit S., et al. Cytotoxic naphthoquinone and a new succinate ester from the soil fungus Fusarium solani PSU-RSPG227. Phytochemistry Lett. 2015;11:106110. doi: 10.1016/j.phytol.2014.11.018. [DOI] [Google Scholar]
- 43.Kakeya H., Kageyama S.-I., Nie L., Onose R., Okada G., Beppu T., et al. Lucilactaene, a new cell cycle inhibitor in p53-transfected cancer cells, produced by a Fusarium sp. J Antibiot (Tokyo) 2001;54:850–854. doi: 10.7164/antibiotics.54.850. [DOI] [PubMed] [Google Scholar]
- 44.Renner M.K., Jensen P.R., Fenical W. Mangicols: structures and biosynthesis of a new class of sesterterpene polyols from a marine fungus of the genus Fusarium. J Org Chem. 2000;65(16):4843–4852. doi: 10.1021/jo000081h. [DOI] [PubMed] [Google Scholar]
- 45.Bogner C.W., Kamdem R.S., Sichtermann G., Matthäus C., Hölscher D., Popp J., et al. Bioactive secondary metabolites with multiple activities from a fungal endophyte. Microb Biotechnol. 2017;10:175–188. doi: 10.1111/1751-7915.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadeem M., Ram M., Alam P., Ahmad M.M., Mohammad A., Al-Qurainy F., et al. Fusarium solani, P1, a new endophytic podophyllotoxin-producing fungus from roots of Podophyllum hexandrum. Afr J Microbiol Res. 2012;6:2493–2499. doi: 10.5897/AJMR11.1596. [DOI] [Google Scholar]
- 47.Hidayat I., Radiastuti N., Rahayu G., Achmadi S., Okane I. Three quinine and cinchonidine producing Fusarium species from Indonesia. Curr Res Environ Appl Mycol. 2016;6:20–34. doi: 10.5943/cream/6/1/3. [DOI] [Google Scholar]
- 48.Cui Y., Yi D., Bai X., Sun B., Zhao Y., Zhang Y. Ginkgolide B produced endophytic fungus (Fusarium oxysporum) isolated from Ginkgo biloba. Fitoterapia. 2012;83:913–920. doi: 10.1016/j.fitote.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Scott P. Recent research on fumonisins: a review. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2012;29:242–248. doi: 10.1080/19440049.2010.546000. [DOI] [PubMed] [Google Scholar]
- 50.Tsavkelova E.A., Bömke C., Netrusov A.I., Weiner J., Tudzynski B. Production of gibberellic acids by an orchid-associated Fusarium proliferatum strain. Fungal Genet Biol. 2008;45:1393–1403. doi: 10.1016/j.fgb.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y.C., Chang H.S., Peng C.F., Lin C.H., Chen I.S. Secondary metabolites from the unripe pulp of Persea americana and their antimycobacterial activities. Food Chem. 2012;135:2904–2909. doi: 10.1016/j.foodchem.2012.07.073. [DOI] [PubMed] [Google Scholar]
- 52.Loizzo M., Tundis R., Menichini F., Saab A., Statti G., Menichini F. Antiproliferative effects of essential oils and their major constituents in human renal adenocarcinoma and amelanotic melanoma cells. Cell Prolif. 2008;41:1002–1012. doi: 10.1111/j.1365-2184.2008.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang L.C., Chiang W., Chang M.Y., Ng L.T., Lin C.C. Antileukemic activity of selected natural products in Taiwan. Am J Chin Med. 2003;31:37–46. doi: 10.1142/S0192415X03000825. [DOI] [PubMed] [Google Scholar]
- 54.Brodmann J., Twele R., Francke W., Yi-Bo L., Xi-qiang S., Ayasse M. Orchid mimics honey bee alarm pheromone in order to attract hornets for pollination. Curr Biol. 2009;19:1368–1372. doi: 10.1016/j.cub.2009.06.067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.